The mosaic structure of the symbiotic plasmid of Rhizobium etli CFN42 and its relation to other symbiotic genome compartments
- PMID: 12801410
- PMCID: PMC193615
- DOI: 10.1186/gb-2003-4-6-r36
The mosaic structure of the symbiotic plasmid of Rhizobium etli CFN42 and its relation to other symbiotic genome compartments
Abstract
Background: Symbiotic bacteria known as rhizobia interact with the roots of legumes and induce the formation of nitrogen-fixing nodules. In rhizobia, essential genes for symbiosis are compartmentalized either in symbiotic plasmids or in chromosomal symbiotic islands. To understand the structure and evolution of the symbiotic genome compartments (SGCs), it is necessary to analyze their common genetic content and organization as well as to study their differences. To date, five SGCs belonging to distinct species of rhizobia have been entirely sequenced. We report the complete sequence of the symbiotic plasmid of Rhizobium etli CFN42, a microsymbiont of beans, and a comparison with other SGC sequences available.
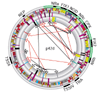
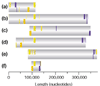
Results: The symbiotic plasmid is a circular molecule of 371,255 base-pairs containing 359 coding sequences. Nodulation and nitrogen-fixation genes common to other rhizobia are clustered in a region of 125 kilobases. Numerous sequences related to mobile elements are scattered throughout. In some cases the mobile elements flank blocks of functionally related sequences, thereby suggesting a role in transposition. The plasmid contains 12 reiterated DNA families that are likely to participate in genomic rearrangements. Comparisons between this plasmid and complete rhizobial genomes and symbiotic compartments already sequenced show a general lack of synteny and colinearity, with the exception of some transcriptional units. There are only 20 symbiotic genes that are shared by all SGCs.
Conclusions: Our data support the notion that the symbiotic compartments of rhizobia genomes are mosaic structures that have been frequently tailored by recombination, horizontal transfer and transposition.
Figures



References
-
- Garrity GM, Johnson KL, Bell JA, Searles DB. In Bergey's Manual of Systematic Bacteriology. New York: Springer-Verlag; 2002. Taxonomic outline of the Procaryotes. Release 3.0.
-
- Kaneko T, Nakamura Y, Sato S, Asamizu E, Kato T, Sasamoto S, Watanabe A, Idesawa K, Ishikawa A, Kawashima K, et al. Complete genome structure of the nitrogen-fixing symbiotic bacterium Mesorhizobium loti. DNA Res. 2000;7:331–338. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

