Characterization of the iron-regulated desA promoter of Streptomyces pilosus as a system for controlled gene expression in actinomycetes
- PMID: 12801423
- PMCID: PMC161790
- DOI: 10.1186/1475-2859-2-5
Characterization of the iron-regulated desA promoter of Streptomyces pilosus as a system for controlled gene expression in actinomycetes
Abstract
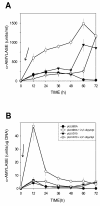
BACKGROUND: The bioavailability of iron is quite low since it is usually present as insoluble complexes. To solve the bioavailability problem microorganisms have developed highly efficient iron-scavenging systems based on the synthesis of siderophores that have high iron affinity. The systems of iron assimilation in microorganisms are strictly regulated to control the intracellular iron levels since at high concentrations iron is toxic for cells. Streptomyces pilosus synthesizes the siderofore desferrioxamine B. The first step in desferrioxamine biosynthesis is decarboxylation of L-lysine to form cadaverine, a desferrioxamine B precursor. This reaction is catalyzed by the lysine decarboxylase, an enzyme encoded by the desA gene that is repressed by iron. RESULTS: The binding of the DmdR (acronym for divalent metal dependent repressor) to the desA promoter in presence of Fe2+ or other divalent ions has been characterized. A 51 bp DNA fragment of the desA promoter containing the 9 bp inverted repeat was sufficient for binding of the DmdR repressor, as observed by the electrophoretic mobility shift assay. The desA mobility shift was prevented by neutralizing DmdR with anti-DmdR antibodies or by chelating the divalent metal in the binding reaction with 2,2'-dipyridyl. Binding to the desA promoter was observed with purified DmdR repressors of Streptomyces coelicolor or Rhodococcus fascians suggesting that there is a common mechanism of iron-regulation in actinomycetes. The complete desA promoter region was coupled using transcriptional fusions to the amy reporter gene (encoding alpha-amylase) in low copy or multicopy Streptomyces vectors. The iron-regulated desA promoter was induced by addition of the iron chelating agent 2,2'-dipyridyl resulting in a strong expression of the reporter gene. CONCLUSIONS: The iron-regulated desA promoter can be used for inducible expression of genes in Streptomyces species, as shown by de-repression of the promoter when coupled to a reporter gene.
Figures
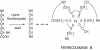



References
-
- Weinberg ED. The development of awareness of iron-withholding defense. Prospect Biol Med. 1993;36:215–221. - PubMed
-
- Neilands JB. Microbial envelope proteins related to iron. Annu Rev Microbiol. 1982;36:285–309. - PubMed
-
- Neilands JB. Siderophores: structure and function of microbial iron transport compounds. J Biol Chem. 1995;270:26723–26726. - PubMed
LinkOut - more resources
Full Text Sources

