A multistep mechanism for the activation of rearrangement in the immune system
- PMID: 12802019
- PMCID: PMC164625
- DOI: 10.1073/pnas.0932635100
A multistep mechanism for the activation of rearrangement in the immune system
Abstract
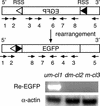
Rearrangement of immune receptor loci is a developmentally controlled process that takes place exclusively in lymphoid cells. We have used a stable transfection system in pre-B cells to show that DNA methylation brings about histone underacetylation, histone H3(K9) methylation, DNaseI resistance, and strong inhibition of both transcription and recombination. Strikingly, this repression is maintained in dividing cells even after removal of the original methyl groups responsible for its establishment, but in this state, rearrangement can now be induced by reacetylation of local histones using the drug Trichostatin A. This same combination of demethylation and histone acetylation is also required to activate germline transcription and recombination from the endogenous kappa locus in vivo. These results indicate that the regulation of rearrangement is carried out by a multilayered synergistic process.
Figures
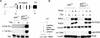




Similar articles
-
A role for histone acetylation in the developmental regulation of VDJ recombination.Science. 2000 Jan 21;287(5452):495-8. doi: 10.1126/science.287.5452.495. Science. 2000. PMID: 10642553
-
Chromatin remodeling directly activates V(D)J recombination.Proc Natl Acad Sci U S A. 1999 Sep 14;96(19):10788-93. doi: 10.1073/pnas.96.19.10788. Proc Natl Acad Sci U S A. 1999. PMID: 10485904 Free PMC article.
-
Chromatin remodeling at the Ig loci prior to V(D)J recombination.J Immunol. 2001 Jul 15;167(2):866-74. doi: 10.4049/jimmunol.167.2.866. J Immunol. 2001. PMID: 11441093
-
Control of chromatin accessibility for V(D)J recombination by interleukin-7.J Leukoc Biol. 2001 Jun;69(6):907-11. J Leukoc Biol. 2001. PMID: 11404375 Review.
-
Epigenetic regulation of antigen receptor rearrangement.Clin Immunol. 2003 Oct;109(1):29-36. doi: 10.1016/s1521-6616(03)00199-2. Clin Immunol. 2003. PMID: 14585273 Review.
Cited by
-
Asynchronous Replication Timing: A Mechanism for Monoallelic Choice During Development.Front Cell Dev Biol. 2021 Oct 1;9:737681. doi: 10.3389/fcell.2021.737681. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34660595 Free PMC article. Review.
-
B cell-specific loss of histone 3 lysine 9 methylation in the V(H) locus depends on Pax5.Nat Immunol. 2004 Aug;5(8):853-61. doi: 10.1038/ni1099. Epub 2004 Jul 18. Nat Immunol. 2004. PMID: 15258579 Free PMC article.
-
Epigenetic regulation of monoallelic rearrangement (allelic exclusion) of antigen receptor genes.Front Immunol. 2014 Dec 5;5:625. doi: 10.3389/fimmu.2014.00625. eCollection 2014. Front Immunol. 2014. PMID: 25538709 Free PMC article. Review.
-
MicroRNA-376b-3p targets RGS1 mRNA to inhibit proliferation, metastasis, and apoptosis in osteosarcoma.Ann Transl Med. 2021 Nov;9(22):1652. doi: 10.21037/atm-21-4949. Ann Transl Med. 2021. PMID: 34988161 Free PMC article.
-
Immunoglobulin kappa enhancers are differentially regulated at the level of chromatin structure.Mol Immunol. 2007 Jul;44(13):3407-15. doi: 10.1016/j.molimm.2007.02.010. Epub 2007 Mar 26. Mol Immunol. 2007. PMID: 17382392 Free PMC article.
References
-
- Schatz, D. G., Oettinger, M. A. & Baltimore, D. (1989) Cell 59, 1035–1048. - PubMed
-
- Oettinger, M. A., Schatz, D. G., Gorka, C. & Baltimore, D. (1990) Science 248, 1517–1523. - PubMed
-
- Gellert, M. (1996) Genes Cells 1, 269–275. - PubMed
-
- Lewis, S. A. (1994) Adv. Immunol. 56, 27–150. - PubMed
-
- Bergman, Y. & Mostoslavsky, R. (1998) Biol. Chem. 379, 401–407. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

