TSG101 interaction with HRS mediates endosomal trafficking and receptor down-regulation
- PMID: 12802020
- PMCID: PMC164637
- DOI: 10.1073/pnas.0932599100
TSG101 interaction with HRS mediates endosomal trafficking and receptor down-regulation
Abstract
Down-regulation of mitogenic signaling in mammalian cells relies in part on endosomal trafficking of activated receptors into lysosomes, where the receptors are degraded. These events are mediated by ubiquitination of the endosomal cargo and its consequent sorting into multivesicular bodies that form at the surfaces of late endosomes. Tumor susceptibility gene 101 (tsg101) recently was found to be centrally involved in this process. Here we report that TSG101 interacts with hepatocyte growth factor-regulated tyrosine kinase substrate (HRS), an early endosomal protein, and that disruption of this interaction impedes endosomal trafficking and endocytosis-mediated degradation of mitogenic receptors. TSG101/HRS interaction occurs between a ubiquitin-binding domain of TSG101 and two distinct proline-rich regions of HRS, and is modulated by a C-terminal TSG101 sequence that resembles a motif targeted in HRS. Mutational perturbation of TSG101/HRS interaction prevented delivery of epidermal growth factor receptor (EGFR) to late endosomes, resulted in the cellular accumulation of ubiquitinated EGFR in early endosomes, and inhibited ligand-induced down-regulation of EGFR. Our results reveal the TSG101 interaction with HRS as a crucial step in endocytic down-regulation of mitogenic signaling and suggest a role for this interaction in linking the functions of early and late endosomes.
Figures


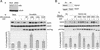

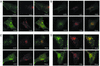
References
-
- Hunter, T. (2000) Cell 100, 113–127. - PubMed
-
- Schlessinger, J. (2000) Cell 103, 211–225. - PubMed
-
- Blume-Jensen, P. & Hunter, T. (2001) Nature 411, 355–365. - PubMed
-
- Di Fiore, P. P. & De Camilli, P. (2001) Cell 106, 1–4. - PubMed
-
- Sorkin, A. & Von Zastrow, M. (2002) Nat. Rev. Mol. Cell Biol. 3, 600–614. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

