Constriction and Dnm1p recruitment are distinct processes in mitochondrial fission
- PMID: 12802067
- PMCID: PMC165089
- DOI: 10.1091/mbc.e02-10-0657
Constriction and Dnm1p recruitment are distinct processes in mitochondrial fission
Abstract
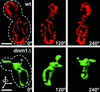
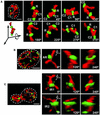
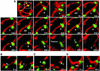
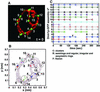
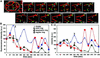
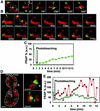
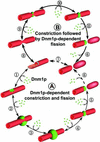
Mitochondria undergo cycles of fusion and fission crucial for organelle homeostasis. Fission is regulated partially by recruitment of the large GTPase Dnm1p to the outer mitochondrial membrane. Using three-dimensional time-lapse fluorescence imaging of Saccharomyces cerevisiae cells, we found that Dnm1p-EGFP appears and disappears at "hot spots" along mitochondrial tubes. It forms patches that convert rapidly into different shapes regardless of whether mitochondrial fission ensues or not. Moreover, the thickness of the mitochondrial matrix displays frequent temporal fluctuations apparently unrelated to fission or to recruitment of Dnm1p-EGFP. These results suggest that mitochondrial fission requires coordination of at least two distinct processes.
Figures


References
-
- Agard, D.A., Hiaroka, Y., Shaw, P., and Sedat, J.W. (1989). Fluorescence microscopy in three dimensions. Methods Cell Biol. 30, 353–377. - PubMed
-
- Hoffmann, H.P., and Avers, C.J. (1973). Mitochondrion of yeast: ultrastructural evidence for one giant, branched organelle per cell. Science 181, 749–751. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

