Three-dimensional structure of the truncated core of the Saccharomyces cerevisiae pyruvate dehydrogenase complex determined from negative stain and cryoelectron microscopy images
- PMID: 1280269
- PMCID: PMC4167662
Three-dimensional structure of the truncated core of the Saccharomyces cerevisiae pyruvate dehydrogenase complex determined from negative stain and cryoelectron microscopy images
Abstract
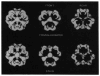
Dihydrolipoamide acyltransferase (E2), a catalytic and structural component of the three functional classes of multienzyme complexes that catalyze the oxidative decarboxylation of alpha-keto acids, forms the central core to which the other components are attached. We have imaged by negative stain and cryoelectron microscopy the truncated dihydrolipoamide acetyltransferase core (60 subunits; M(r) = 2.7 x 10(6)) of the Saccharomyces cerevisiae pyruvate dehydrogenase complex. Using icosahedral particle reconstruction techniques, we determined its structure to 25 A resolution. Although the model derived from the negative stain reconstruction was approximately 20% smaller than the model derived from the frozen-hydrated data, when corrected for the effects of the electron microscope contrast transfer functions, the reconstructions showed excellent correspondence. The pentagonal dodecahedron-shaped macromolecule has a maximum diameter, as measured along the 3-fold axis, of approximately 226 A (frozen-hydrated value), and 12 large openings (approximately 63 A in diameter) on the 5-fold axes that lead into a large solvent-accessible cavity (approximately 76-140 A diameter). The 20 vertices consist of cone-shaped trimers, each with a flattened base on the outside of the structure and an apex directed toward the center. The trimers are interconnected by 20 A thick "bridges" on the 2-fold axes. These studies also show that the highest resolution features apparent in the frozen-hydrated reconstruction are revealed in a filtered reconstruction of the stained molecule.
Figures





Similar articles
-
Direct evidence for the size and conformational variability of the pyruvate dehydrogenase complex revealed by three-dimensional electron microscopy. The "breathing" core and its functional relationship to protein dynamics.J Biol Chem. 2001 Jun 15;276(24):21704-13. doi: 10.1074/jbc.M101765200. Epub 2001 Apr 2. J Biol Chem. 2001. PMID: 11285267
-
On the unique structural organization of the Saccharomyces cerevisiae pyruvate dehydrogenase complex.J Biol Chem. 1997 Feb 28;272(9):5757-64. doi: 10.1074/jbc.272.9.5757. J Biol Chem. 1997. PMID: 9038189 Free PMC article.
-
Cryoelectron microscopy of mammalian pyruvate dehydrogenase complex.J Biol Chem. 1991 Dec 25;266(36):24650-6. J Biol Chem. 1991. PMID: 1761562
-
The pyruvate dehydrogenase multi-enzyme complex from Gram-negative bacteria.Biochim Biophys Acta. 1998 Jun 29;1385(2):353-66. doi: 10.1016/s0167-4838(98)00079-x. Biochim Biophys Acta. 1998. PMID: 9655933 Review.
-
Structure-function relationships in dihydrolipoamide acyltransferases.J Biol Chem. 1990 Jun 5;265(16):8971-4. J Biol Chem. 1990. PMID: 2188967 Review. No abstract available.
Cited by
-
Three-dimensional structure of the human herpesvirus 8 capsid.J Virol. 2000 Oct;74(20):9646-54. doi: 10.1128/jvi.74.20.9646-9654.2000. J Virol. 2000. PMID: 11000237 Free PMC article.
-
Solution structure and characterisation of the human pyruvate dehydrogenase complex core assembly.J Mol Biol. 2010 May 28;399(1):71-93. doi: 10.1016/j.jmb.2010.03.043. Epub 2010 Mar 31. J Mol Biol. 2010. PMID: 20361979 Free PMC article.
-
Rearrangement of the 16S precursor subunits is essential for the formation of the active 20S proteasome.Biophys J. 2004 Dec;87(6):4098-105. doi: 10.1529/biophysj.104.051144. Epub 2004 Sep 10. Biophys J. 2004. PMID: 15361411 Free PMC article.
-
Structures of the human pyruvate dehydrogenase complex cores: a highly conserved catalytic center with flexible N-terminal domains.Structure. 2008 Jan;16(1):104-14. doi: 10.1016/j.str.2007.10.024. Structure. 2008. PMID: 18184588 Free PMC article.
-
"Scanning mutagenesis" of the amino acid sequences flanking phosphorylation site 1 of the mitochondrial pyruvate dehydrogenase complex.Front Plant Sci. 2012 Jul 16;3:153. doi: 10.3389/fpls.2012.00153. eCollection 2012. Front Plant Sci. 2012. PMID: 22811682 Free PMC article.
References
-
- Perham RN. Biochemistry. 1991;30:8501–8512. - PubMed
-
- Reed LJ, Hackert ML. J Biol Chem. 1990;265:8971–8974. - PubMed
-
- Guest JR, Angier SJ, Russell GC. Ann N Y Acad Sci. 1989;573:76–99. - PubMed
-
- Gopalakrishnan S, Rahmatullah M, Radke GA, Powers-Greenwood S, Roche TE. Biochem Biophys Res Commun. 1989;160:715–721. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

