Tic40, a membrane-anchored co-chaperone homolog in the chloroplast protein translocon
- PMID: 12805212
- PMCID: PMC162133
- DOI: 10.1093/emboj/cdg281
Tic40, a membrane-anchored co-chaperone homolog in the chloroplast protein translocon
Abstract
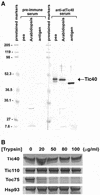
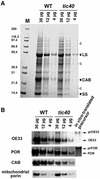
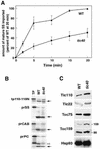
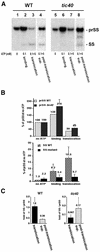
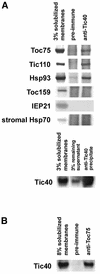
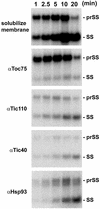
The function of Tic40 during chloroplast protein import was investigated. Tic40 is an inner envelope membrane protein with a large hydrophilic domain located in the stroma. Arabidopsis null mutants of the atTic40 gene were very pale green and grew slowly but were not seedling lethal. Isolated mutant chloroplasts imported precursor proteins at a lower rate than wild-type chloroplasts. Mutant chloroplasts were normal in allowing binding of precursor proteins. However, during subsequent translocation across the inner membrane, fewer precursors were translocated and more precursors were released from the mutant chloroplasts. Cross-linking experiments demonstrated that Tic40 was part of the translocon complex and functioned at the same stage of import as Tic110 and Hsp93, a member of the Hsp100 family of molecular chaperones. Tertiary structure prediction and immunological studies indicated that the C-terminal portion of Tic40 contains a TPR domain followed by a domain with sequence similarity to co-chaperones Sti1p/Hop and Hip. We propose that Tic40 functions as a co-chaperone in the stromal chaperone complex that facilitates protein translocation across the inner membrane.
Figures



References
-
- Bauerle C., Dorl,J. and Keegstra,K. (1991) Kinetic analysis of the transport of thylakoid lumenal proteins in experiments using intact chloroplasts. J. Biol. Chem., 266, 5884–5890. - PubMed
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

