Detailed map of a cis-regulatory input function
- PMID: 12805558
- PMCID: PMC164651
- DOI: 10.1073/pnas.1230759100
Detailed map of a cis-regulatory input function
Abstract
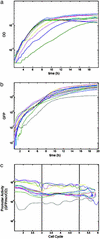
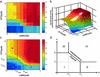
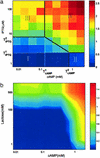
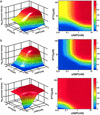
Most genes are regulated by multiple transcription factors that bind specific sites in DNA regulatory regions. These cis-regulatory regions perform a computation: the rate of transcription is a function of the active concentrations of each of the input transcription factors. Here, we used accurate gene expression measurements from living cell cultures, bearing GFP reporters, to map in detail the input function of the classic lacZYA operon of Escherichia coli, as a function of about a hundred combinations of its two inducers, cAMP and isopropyl beta-d-thiogalactoside (IPTG). We found an unexpectedly intricate function with four plateau levels and four thresholds. This result compares well with a mathematical model of the binding of the regulatory proteins cAMP receptor protein (CRP) and LacI to the lac regulatory region. The model is also used to demonstrate that with few mutations, the same region could encode much purer AND-like or even OR-like functions. This possibility means that the wild-type region is selected to perform an elaborate computation in setting the transcription rate. The present approach can be generally used to map the input functions of other genes.
Figures

References
-
- Jacob, F. & Monod, J. (1961) J. Mol. Biol. 3, 318-356. - PubMed
-
- Beckwith, J. & Zipser, D., eds. (1970) The Lactose Operon (Cold Spring Harbor Lab. Press, Plainview, NY).
-
- Muller-Hill, B. (1996) The lac Operon (de Gruyter, Berlin).
-
- Savageau, M. A. (1976) Biochemical Systems Analysis: A Study of Function and Design in Molecular Biology (Addison–Wesley, Reading, MA).
-
- Hartwell, L. H., Hopfield, J. J., Leibler, S. & Murray, A. W. (1999) Nature 402, C47-C52. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

