Expansion of the receptor-like kinase/Pelle gene family and receptor-like proteins in Arabidopsis
- PMID: 12805585
- PMCID: PMC166995
- DOI: 10.1104/pp.103.021964
Expansion of the receptor-like kinase/Pelle gene family and receptor-like proteins in Arabidopsis
Abstract
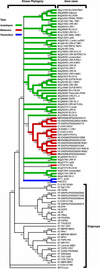
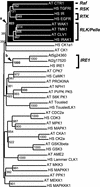
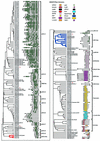
Receptor-like kinases (RLKs) are a family of transmembrane proteins with versatile N-terminal extracellular domains and C-terminal intracellular kinases. They control a wide range of physiological responses in plants and belong to one of the largest gene families in the Arabidopsis genome with more than 600 members. Interestingly, this gene family constitutes 60% of all kinases in Arabidopsis and accounts for nearly all transmembrane kinases in Arabidopsis. Analysis of four fungal, six metazoan, and two Plasmodium sp. genomes indicates that the family was represented in all but fungal genomes, indicating an ancient origin for the family with a more recent expansion only in the plant lineages. The RLK/Pelle family can be divided into several subfamilies based on three independent criteria: the phylogeny based on kinase domain sequences, the extracellular domain identities, and intron locations and phases. A large number of receptor-like proteins (RLPs) resembling the extracellular domains of RLKs are also found in the Arabidopsis genome. However, not all RLK subfamilies have corresponding RLPs. Several RLK/Pelle subfamilies have undergone differential expansions. More than 33% of the RLK/Pelle members are found in tandem clusters, substantially higher than the genome average. In addition, 470 of the RLK/Pelle family members are located within the segmentally duplicated regions in the Arabidopsis genome and 268 of them have a close relative in the corresponding regions. Therefore, tandem duplications and segmental/whole-genome duplications represent two of the major mechanisms for the expansion of the RLK/Pelle family in Arabidopsis.
Figures


References
-
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 - PubMed
-
- Becraft PW (1998) Receptor kinases in plant development. Trends Plant Sci 3: 384–388
-
- Bleecker AB, Kende H (2000) Ethylene: a gaseous signal molecule in plants. Annu Rev Cell Dev Biol 16: 1–18 - PubMed
-
- Brand U, Fletcher JC, Hobe M, Meyerowitz EM, Simon R (2000) Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289: 617–619 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

