Microarray analysis of the nitrate response in Arabidopsis roots and shoots reveals over 1,000 rapidly responding genes and new linkages to glucose, trehalose-6-phosphate, iron, and sulfate metabolism
- PMID: 12805587
- PMCID: PMC166997
- DOI: 10.1104/pp.103.021253
Microarray analysis of the nitrate response in Arabidopsis roots and shoots reveals over 1,000 rapidly responding genes and new linkages to glucose, trehalose-6-phosphate, iron, and sulfate metabolism
Abstract
The genomic response to low levels of nitrate was studied in Arabidopsis using the Affymetrix ATH1 chip containing more than 22,500 probe sets. Arabidopsis plants were grown hydroponically in sterile liquid culture on ammonium as the sole source of nitrogen for 10 d, then treated with 250 microm nitrate for 20 min. The response to nitrate was much stronger in roots (1,176 genes showing increased or decreased mRNA levels) than in shoots (183 responding genes). In addition to known nitrate-responsive genes (e.g. those encoding nitrate transporters, nitrate reductase, nitrite reductase, ferredoxin reductase, and enzymes in the pentose phosphate pathway), genes encoding novel metabolic and potential regulatory proteins were found. These genes encode enzymes in glycolysis (glucose-6-phosphate isomerase and phosphoglycerate mutase), in trehalose-6-P metabolism (trehalose-6-P synthase and trehalose-6-P phosphatase), in iron transport/metabolism (nicotianamine synthase), and in sulfate uptake/reduction. In many cases, only a few select genes out of several in small gene families were induced by nitrate. These results show that the effect of nitrate on gene expression is substantial (affecting almost 10% of the genes with detectable mRNA levels) yet selective and affects many genes involved in carbon and nutrient metabolism.
Figures

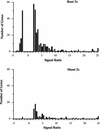





References
-
- Crawford NM, Glass ADM (1998) Molecular and physiological aspects of nitrate uptake in plants. Trends Plant Sci 3: 389–395
-
- Crawford NM, Kahn ML, Leustek T, Long SR (2000) Nitrogen and sulfur. In B Buchanan, W Gruissem, RL Jones, eds, Biochemistry and Molecular Biology of Plants. American Society of Plant Physiologists, Rockville, MD, pp 786–849
-
- Dennis DT, Blakeley SD (2000) Carbohydrate metabolism. In B Buchanan, W Gruissem, RL Jones, eds, Biochemistry and Molecular Biology of Plants. American Society of Plant Physiologists, Rockville, MD, pp 630–675
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

