Identification of glycosylphosphatidylinositol-anchored proteins in Arabidopsis. A proteomic and genomic analysis
- PMID: 12805588
- PMCID: PMC166998
- DOI: 10.1104/pp.103.021170
Identification of glycosylphosphatidylinositol-anchored proteins in Arabidopsis. A proteomic and genomic analysis
Abstract
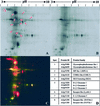
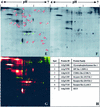
In a recent bioinformatic analysis, we predicted the presence of multiple families of cell surface glycosylphosphatidylinositol (GPI)-anchored proteins (GAPs) in Arabidopsis (G.H.H. Borner, D.J. Sherrier, T.J. Stevens, I.T. Arkin, P. Dupree [2002] Plant Physiol 129: 486-499). A number of publications have since demonstrated the importance of predicted GAPs in diverse physiological processes including root development, cell wall integrity, and adhesion. However, direct experimental evidence for their GPI anchoring is mostly lacking. Here, we present the first, to our knowledge, large-scale proteomic identification of plant GAPs. Triton X-114 phase partitioning and sensitivity to phosphatidylinositol-specific phospholipase C were used to prepare GAP-rich fractions from Arabidopsis callus cells. Two-dimensional fluorescence difference gel electrophoresis and one-dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis demonstrated the existence of a large number of phospholipase C-sensitive Arabidopsis proteins. Using liquid chromatography-tandem mass spectrometry, 30 GAPs were identified, including six beta-1,3 glucanases, five phytocyanins, four fasciclin-like arabinogalactan proteins, four receptor-like proteins, two Hedgehog-interacting-like proteins, two putative glycerophosphodiesterases, a lipid transfer-like protein, a COBRA-like protein, SKU5, and SKS1. These results validate our previous bioinformatic analysis of the Arabidopsis protein database. Using the confirmed GAPs from the proteomic analysis to train the search algorithm, as well as improved genomic annotation, an updated in silico screen yielded 64 new candidates, raising the total to 248 predicted GAPs in Arabidopsis.
Figures
References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 - PubMed
-
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 - PubMed
-
- Bateman A, Bycroft M (2000) The structure of a LysM domain from E. coli membrane-bound lytic murein transglycosylase D (MltD). J Mol Biol 299: 1113–1119 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

