Genomic comparison of P-type ATPase ion pumps in Arabidopsis and rice
- PMID: 12805592
- PMCID: PMC167002
- DOI: 10.1104/pp.103.021923
Genomic comparison of P-type ATPase ion pumps in Arabidopsis and rice
Erratum in
- Plant Physiol. 2005 Jul;138(3):1807
Abstract
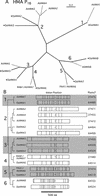
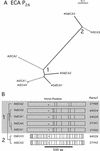
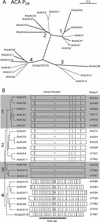
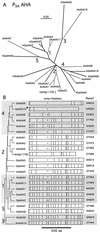
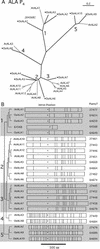
Members of the P-type ATPase ion pump superfamily are found in all three branches of life. Forty-six P-type ATPase genes were identified in Arabidopsis, the largest number yet identified in any organism. The recent completion of two draft sequences of the rice (Oryza sativa) genome allows for comparison of the full complement of P-type ATPases in two different plant species. Here, we identify a similar number (43) in rice, despite the rice genome being more than three times the size of Arabidopsis. The similarly large families suggest that both dicots and monocots have evolved with a large preexisting repertoire of P-type ATPases. Both Arabidopsis and rice have representative members in all five major subfamilies of P-type ATPases: heavy-metal ATPases (P1B), Ca2+-ATPases (endoplasmic reticulum-type Ca2+-ATPase and autoinhibited Ca2+-ATPase, P2A and P2B), H+-ATPases (autoinhibited H+-ATPase, P3A), putative aminophospholipid ATPases (ALA, P4), and a branch with unknown specificity (P5). The close pairing of similar isoforms in rice and Arabidopsis suggests potential orthologous relationships for all 43 rice P-type ATPases. A phylogenetic comparison of protein sequences and intron positions indicates that the common angiosperm ancestor had at least 23 P-type ATPases. Although little is known about unique and common features of related pumps, clear differences between some members of the calcium pumps indicate that evolutionarily conserved clusters may distinguish pumps with either different subcellular locations or biochemical functions.
Figures


References
-
- Altschul S, Gish W, Miller W, Myers E, Lipman D (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 - PubMed
-
- Arango M, Gévaudant F, Oufattole M, Boutry M (2003) The plasma membrane proton pump ATPase: the significance of gene subfamilies. Planta 216: 355–365 - PubMed
-
- Axelsen KB, Palmgren MG (1998) Evolution of substrate specificities in the P-type ATPase superfamily. J Mol Evol 46: 84–101 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

