Arabidopsis genes involved in acyl lipid metabolism. A 2003 census of the candidates, a study of the distribution of expressed sequence tags in organs, and a web-based database
- PMID: 12805597
- PMCID: PMC167007
- DOI: 10.1104/pp.103.022988
Arabidopsis genes involved in acyl lipid metabolism. A 2003 census of the candidates, a study of the distribution of expressed sequence tags in organs, and a web-based database
Abstract
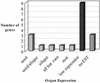
The genome of Arabidopsis has been searched for sequences of genes involved in acyl lipid metabolism. Over 600 encoded proteins have been identified, cataloged, and classified according to predicted function, subcellular location, and alternative splicing. At least one-third of these proteins were previously annotated as "unknown function" or with functions unrelated to acyl lipid metabolism; therefore, this study has improved the annotation of over 200 genes. In particular, annotation of the lipolytic enzyme group (at least 110 members total) has been improved by the critical examination of the biochemical literature and the sequences of the numerous proteins annotated as "lipases." In addition, expressed sequence tag (EST) data have been surveyed, and more than 3,700 ESTs associated with the genes were cataloged. Statistical analysis of the number of ESTs associated with specific cDNA libraries has allowed calculation of probabilities of differential expression between different organs. More than 130 genes have been identified with a statistical probability > 0.95 of preferential expression in seed, leaf, root, or flower. All the data are available as a Web-based database, the Arabidopsis Lipid Gene database (http://www.plantbiology.msu.edu/lipids/genesurvey/index.htm). The combination of the data of the Lipid Gene Catalog and the EST analysis can be used to gain insights into differential expression of gene family members and sets of pathway-specific genes, which in turn will guide studies to understand specific functions of individual genes.
Figures

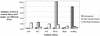
References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 - PubMed
-
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 - PubMed
-
- Arondel V, Vergnolle C, Cantrel C, Kader JC (2000) Lipid transfer proteins are encoded by a small multigene family in Arabidopsis thaliana. Plant Sci 157: 1–12 - PubMed
-
- Audic S, Claverie JM (1997) The significance of digital expression profiles. Genome Res 7: 986–995 - PubMed
-
- Beisson F, Tiss A, Rivière C, Verger R (2000) Methods for lipase detection and assay: a critical review. Eur J Lipid Sci Technol 2: 133–153
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

