AtSig5 is an essential nucleus-encoded Arabidopsis sigma-like factor
- PMID: 12805603
- PMCID: PMC167013
- DOI: 10.1104/pp.102.017913
AtSig5 is an essential nucleus-encoded Arabidopsis sigma-like factor
Abstract
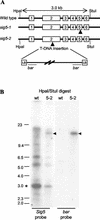
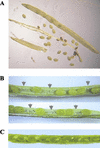
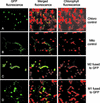
Transcription of chloroplast genes is subject to control by nucleus-encoded proteins. The chloroplast-encoded RNA polymerase (PEP) is a eubacterial-type RNA polymerase that is presumed to assemble with nucleus-encoded sigma-factors mediating promoter recognition. Recently, families of sigma-factor genes have been identified in several plants including Arabidopsis. One of these genes, Arabidopsis SIG5, encodes a sigma-factor, AtSig5, which is phylogenetically distinct from the other family members. To investigate the role of this plant sigma-factor, two different insertional alleles of the SIG5 gene were identified and characterized. Heterozygous mutant plants showed no visible leaf phenotype, but exhibited siliques containing aborted embryos and unfertilized ovules. Our inability to recover plants homozygous for a SIG5 gene disruption indicates that SIG5 is an essential gene. SIG5 transcripts accumulate in flower tissues, consistent with a role for AtSig5 protein in reproduction. Therefore, SIG5 encodes an essential member of the Arabidopsis sigma-factor family that plays a role in plant reproduction in addition to its previously proposed role in leaf chloroplast gene expression.
Figures



References
-
- Allison LA (2000) The role of sigma factors in plastid transcription. Biochimie 82: 537–548 - PubMed
-
- Beardslee T, Chowdhury SR, Chang CC, Jaiswal P, Lerbs-Mache S, Stern D, Allison LA (2002) A nucleus-encoded maize protein with sigma factor activity accumulates in both chloroplasts and mitochondria. Plant J 31: 199–209 - PubMed
-
- Burgess RR, Anthony L (2001) How sigma docks to RNA polymerase and what sigma does. Curr Opin Microbiol 4: 126–131 - PubMed
-
- Campbell EA, Muzzin O, Chlenov M, Sun JL, Olson CA, Weinman O, Trester-Zedlitz ML, Darst SA (2002) Structure of the bacterial RNA polymerase promoter specificity sigma subunit. Mol Cell 9: 527–539 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

