AtCHIP, a U-box-containing E3 ubiquitin ligase, plays a critical role in temperature stress tolerance in Arabidopsis
- PMID: 12805616
- PMCID: PMC167026
- DOI: 10.1104/pp.103.020800
AtCHIP, a U-box-containing E3 ubiquitin ligase, plays a critical role in temperature stress tolerance in Arabidopsis
Abstract
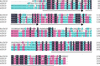
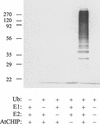
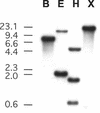
The Arabidopsis gene AtCHIP encodes a protein with three tetratricopeptide repeats and a U-box domain, which is structurally similar to the animal CHIP proteins, a new class of E3 ubiquitin ligases. Like animal CHIP proteins, AtCHIP has E3 ubiquitin ligase activity in vitro. AtCHIP is a single-copy gene, and its transcript is up-regulated by several stress conditions such as low and high temperatures. However, increased AtCHIP expression alone was not correlated with increased stress tolerance; in fact, overexpression of AtCHIP in Arabidopsis rendered plants more sensitive to both low- and high-temperature treatments. Higher electrolyte leakage was observed in leaves of AtCHIP overexpression plants after chilling temperature treatment, suggesting that membrane function is likely impaired in these plants under such a condition. These results indicate that AtCHIP plays an important role in plant cellular metabolism under temperature stress conditions.
Figures







Similar articles
-
AtCHIP functions as an E3 ubiquitin ligase of protein phosphatase 2A subunits and alters plant response to abscisic acid treatment.Plant J. 2006 May;46(4):649-57. doi: 10.1111/j.1365-313X.2006.02730.x. Plant J. 2006. PMID: 16640601
-
The chloroplast protease subunit ClpP4 is a substrate of the E3 ligase AtCHIP and plays an important role in chloroplast function.Plant J. 2007 Jan;49(2):228-37. doi: 10.1111/j.1365-313X.2006.02963.x. Plant J. 2007. PMID: 17241447
-
The E3 ligase AtCHIP positively regulates Clp proteolytic subunit homeostasis.J Exp Bot. 2015 Sep;66(19):5809-20. doi: 10.1093/jxb/erv286. Epub 2015 Jun 17. J Exp Bot. 2015. PMID: 26085677
-
The E3 ligase AtCHIP ubiquitylates FtsH1, a component of the chloroplast FtsH protease, and affects protein degradation in chloroplasts.Plant J. 2007 Oct;52(2):309-21. doi: 10.1111/j.1365-313X.2007.03239.x. Epub 2007 Aug 21. Plant J. 2007. PMID: 17714429
-
ZmRFP1, the putative ortholog of SDIR1, encodes a RING-H2 E3 ubiquitin ligase and responds to drought stress in an ABA-dependent manner in maize.Gene. 2012 Mar 10;495(2):146-53. doi: 10.1016/j.gene.2011.12.028. Epub 2011 Dec 29. Gene. 2012. PMID: 22245611
Cited by
-
Molecular and functional characterization of the only known hemiascomycete ortholog of the carboxyl terminus of Hsc70-interacting protein CHIP in the yeast Yarrowia lipolytica.Cell Stress Chaperones. 2012 Mar;17(2):229-41. doi: 10.1007/s12192-011-0302-6. Epub 2011 Oct 29. Cell Stress Chaperones. 2012. PMID: 22038197 Free PMC article.
-
Expression analysis of genes associated with the induction of the carbon-concentrating mechanism in Chlamydomonas reinhardtii.Plant Physiol. 2008 May;147(1):340-54. doi: 10.1104/pp.107.114652. Epub 2008 Mar 5. Plant Physiol. 2008. PMID: 18322145 Free PMC article.
-
The involvement of wheat F-box protein gene TaFBA1 in the oxidative stress tolerance of plants.PLoS One. 2015 Apr 23;10(4):e0122117. doi: 10.1371/journal.pone.0122117. eCollection 2015. PLoS One. 2015. PMID: 25906259 Free PMC article.
-
The Role of E3 Ubiquitin Ligases in Chloroplast Function.Int J Mol Sci. 2022 Aug 25;23(17):9613. doi: 10.3390/ijms23179613. Int J Mol Sci. 2022. PMID: 36077009 Free PMC article. Review.
-
Protein replenishment in pitcher fluids of Nepenthes × ventrata revealed by quantitative proteomics (SWATH-MS) informed by transcriptomics.J Plant Res. 2019 Sep;132(5):681-694. doi: 10.1007/s10265-019-01130-w. Epub 2019 Aug 17. J Plant Res. 2019. PMID: 31422552
References
-
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 - PubMed
-
- Blatch GL, Lassle M (1999) The tetratricopeptide repeat: a structural motif mediating protein-protein interaction. BioEssays 21: 932–939 - PubMed
-
- Blatch GL, Lassle M, Zetter RB, Kundra V (1997) Isolation of a mouse cDNA encoding mSTI1, a stress inducible protein containing the TPR motif. Gene 194: 277–282 - PubMed
-
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

