Comparative analysis of SET domain proteins in maize and Arabidopsis reveals multiple duplications preceding the divergence of monocots and dicots
- PMID: 12805620
- PMCID: PMC167030
- DOI: 10.1104/pp.102.013722
Comparative analysis of SET domain proteins in maize and Arabidopsis reveals multiple duplications preceding the divergence of monocots and dicots
Abstract
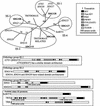
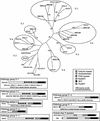
Histone proteins play a central role in chromatin packaging, and modification of histones is associated with chromatin accessibility. SET domain [Su(var)3-9, Enhancer-of-zeste, Trithorax] proteins are one class of proteins that have been implicated in regulating gene expression through histone methylation. The relationships of 22 SET domain proteins from maize (Zea mays) and 32 SET domain proteins from Arabidopsis were evaluated by phylogenetic analysis and domain organization. Our analysis reveals five classes of SET domain proteins in plants that can be further divided into 19 orthology groups. In some cases, such as the Enhancer of zeste-like and trithorax-like proteins, plants and animals contain homologous proteins with a similar organization of domains outside of the SET domain. However, a majority of plant SET domain proteins do not have an animal homolog with similar domain organization, suggesting that plants have unique mechanisms to establish and maintain chromatin states. Although the domains present in plant and animal SET domain proteins often differ, the domains found in the plant proteins have been generally implicated in protein-protein interactions, indicating that most SET domain proteins operate in complexes. Combined analysis of the maize and Arabidopsis SET domain proteins reveals that duplication of SET domain proteins in plants is extensive and has occurred via multiple mechanisms that preceded the divergence of monocots and dicots.
Figures






References
-
- Aasland R, Gibson TJ, Stewart AF (1995) The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem Sci 20: 56-59 - PubMed
-
- Aasland R, Stewart AF, Gibson T (1996) The SANT domain: a putative DNA-binding domain in the SWI-SNF and ADA complexes, the transcriptional co-repressor N-CoR and TFIIIB. Trends Biochem Sci 21: 87-88 - PubMed
-
- Alvarez-Venegas R, Avramova Z (2001) Two Arabidopsis homologs of the animal trithorax genes: a new structural domain is a signature feature of the trithorax gene family. Gene 271: 215-221 - PubMed
-
- Alvarez-Venegas R, Avramova Z (2002) SET-domain proteins of the Su(var)3-9, E(z) and trithorax families. Gene 285: 25-37 - PubMed
-
- Balciunas D, Ronne H (2000) Evidence of domain swapping within the jumonji family of transcription factors. Trends Biochem Sci 25: 274-276 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

