Identification and characterization of aluminum tolerance loci in Arabidopsis (Landsberg erecta x Columbia) by quantitative trait locus mapping. A physiologically simple but genetically complex trait
- PMID: 12805622
- PMCID: PMC167032
- DOI: 10.1104/pp.103.023085
Identification and characterization of aluminum tolerance loci in Arabidopsis (Landsberg erecta x Columbia) by quantitative trait locus mapping. A physiologically simple but genetically complex trait
Abstract
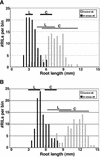
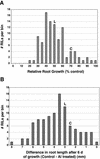
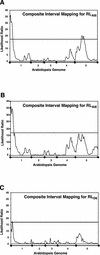
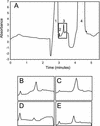
Aluminum (Al) toxicity, which is caused by the solubilization of Al3+ in acid soils resulting in inhibition of root growth and nutrient/water acquisition, is a serious limitation to crop production, because up to one-half of the world's potentially arable land is acidic. To date, however, no Al tolerance genes have yet been cloned. The physiological mechanisms of tolerance are somewhat better understood; the major documented mechanism involves the Al-activated release of Al-binding organic acids from the root tip, preventing uptake into the primary site of toxicity. In this study, a quantitative trait loci analysis of Al tolerance in Arabidopsis was conducted, which also correlated Al tolerance quantitative trait locus (QTL) with physiological mechanisms of tolerance. The analysis identified two major loci, which explain approximately 40% of the variance in Al tolerance observed among recombinant inbred lines derived from Landsberg erecta (sensitive) and Columbia (tolerant). We characterized the mechanism by which tolerance is achieved, and we found that the two QTL cosegregate with an Al-activated release of malate from Arabidopsis roots. Although only two of the QTL have been identified, malate release explains nearly all (95%) of the variation in Al tolerance in this population. Al tolerance in Landsberg erecta x Columbia is more complex genetically than physiologically, in that a number of genes underlie a single physiological mechanism involving root malate release. These findings have set the stage for the subsequent cloning of the genes responsible for the Al tolerance QTL, and a genomics-based cloning strategy and initial progress on this are also discussed.
Figures

References
-
- Alonso-Blanco C, Koornneef M (2000) Naturally occurring variation in Arabidopsis: an underexploited resource for plant genetics. Trends Plant Sci 5: 22-29 - PubMed
-
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796-815 - PubMed
-
- Basten CJ, Weir BS, Zeng ZB (1999) QTL Cartographer. Computer Program Version 1.13. North Carolina State University, Raleigh, NC
-
- Baykov AA, Evtushenko OA, Avaeva SM (1988) A malachite green procedure for orthophosphate determination and its use in alkaline phosphatase-based enzyme immunoassay. Anal Biochem 171: 266-270 - PubMed
-
- Berzonsky WA (1992) The genomic inheritance of Al tolerance in ′Atlas 66′ wheat. Genome 35: 689-693
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

