A role for the GCC-box in jasmonate-mediated activation of the PDF1.2 gene of Arabidopsis
- PMID: 12805630
- PMCID: PMC167040
- DOI: 10.1104/pp.102.017814
A role for the GCC-box in jasmonate-mediated activation of the PDF1.2 gene of Arabidopsis
Abstract
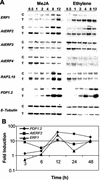
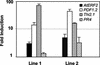
The PDF1.2 gene of Arabidopsis encoding a plant defensin is commonly used as a marker for characterization of the jasmonate-dependent defense responses. Here, using PDF1.2 promoter-deletion lines linked to the beta-glucoronidase-reporter gene, we examined putative promoter elements associated with jasmonate-responsive expression of this gene. Using stably transformed plants, we first characterized the extended promoter region that positively regulates basal expression from the PDF1.2 promoter. Second, using promoter deletion constructs including one from which the GCC-box region was deleted, we observed a substantially lower response to jasmonate than lines carrying this motif. In addition, point mutations introduced into the core GCC-box sequence substantially reduced jasmonate responsiveness, whereas addition of a 20-nucleotide-long promoter element carrying the core GCC-box and flanking nucleotides provided jasmonate responsiveness to a 35S minimal promoter. Taken together, these results indicated that the GCC-box plays a key role in conferring jasmonate responsiveness to the PDF1.2 promoter. However, deletion or specific mutations introduced into the core GCC-box did not completely abolish the jasmonate responsiveness of the promoter, suggesting that the other promoter elements lying downstream from the GCC-box region may also contribute to jasmonate responsiveness. In other experiments, we identified a jasmonate- and pathogen-responsive ethylene response factor transcription factor, AtERF2, which when overexpressed in transgenic Arabidopsis plants activated transcription from the PDF1.2, Thi2.1, and PR4 (basic chitinase) genes, all of which contain a GCC-box sequence in their promoters. Our results suggest that in addition to their roles in regulating ethylene-mediated gene expression, ethylene response factors also appear to play important roles in regulating jasmonate-responsive gene expression, possibly via interaction with the GCC-box.
Figures



References
-
- Berrocal-Lobo M, Molina A, Solano R (2002) Constitutive expression of ETHYLENE-RESPONSE-FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. Plant J 29: 23-32 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

