MTG1 codes for a conserved protein required for mitochondrial translation
- PMID: 12808030
- PMCID: PMC194879
- DOI: 10.1091/mbc.e02-10-0636
MTG1 codes for a conserved protein required for mitochondrial translation
Abstract
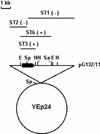
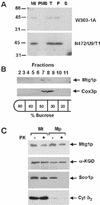
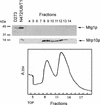
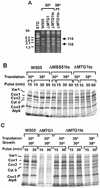
The MTG1 gene of Saccharomyces cerevisiae, corresponding to ORF YMR097c on chromosome XIII, codes for a mitochondrial protein essential for respiratory competence. A human homologue of Mtg1p capable of partially rescuing the respiratory deficiency of a yeast mtg1 mutant has also been localized in mitochondria. Mtg1p is a member of a family of GTPases with largely unknown functions. The respiratory deficiency of mtg1 mutants stems from a defect in mitochondrial protein synthesis. Mutations in the 21S rRNA locus are able to suppress the translation defect of mtg1 null mutants. This points to the 21S rRNA or the large ribosomal subunit as the most likely target of Mtg1p action. The presence of mature size 15S and 21S mitochondrial rRNAs in mtg1 mutants excludes Mtg1p from being involved in transcription or processing of these RNAs. More likely, Mtg1p functions in assembly of the large ribosomal subunit. This is consistent with the peripheral localization of Mtg1p on the matrix side of the inner membrane and the results of in vivo mitochondrial translation assays in a temperature-sensitive mtg1 mutant.
Figures



References
-
- Ban, N., Nisson, P., Hansen, J., Moore, P.B., and Steitz, T.S. (2000). The complete atomic structure of the large ribosomal subunit at 2.4Å resolution. Science 289, 905-920. - PubMed
-
- Bassler, J., Grandi, P., Gadal, O., Lessman, T., Petfalski, E., Tollerway, D., Lechner, J., and Hurt, E. (2001). Identification of a 60S preribosomal particle that is closely linked to nuclear export. Mol. Cell 8, 517-529. - PubMed
-
- Beers, J., Glerum, D.M., and Tzagoloff, A. (1997). Purification, characterization, and localization of yeast Cox17p, a mitochondrial copper shuttle. J. Biol. Chem. 272, 33191-33196. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

