A novel Golgi membrane protein is a partner of the ARF exchange factors Gea1p and Gea2p
- PMID: 12808035
- PMCID: PMC194885
- DOI: 10.1091/mbc.e02-10-0693
A novel Golgi membrane protein is a partner of the ARF exchange factors Gea1p and Gea2p
Abstract
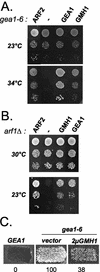
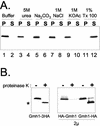
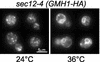
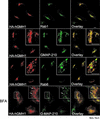
The Sec7 domain guanine nucleotide exchange factors (GEFs) for the GTPase ARF are highly conserved regulators of membrane dynamics and protein trafficking. The interactions of large ARF GEFs with cellular membranes for localization and/or activation are likely to participate in regulated recruitment of ARF and effectors. However, these interactions remain largely unknown. Here we characterize Gmh1p, the first Golgi transmembrane-domain partner of any of the high-molecular-weight ARF-GEFs. Gmh1p is an evolutionarily conserved protein. We demonstrate molecular interaction between the yeast Gmh1p and the large ARF-GEFs Gea1p and Gea2p. This interaction involves a domain of Gea1p and Gea2p that is conserved in the eukaryotic orthologues of the Gea proteins. A single mutation in a conserved amino acid residue of this domain is sufficient to abrogate the interaction, whereas the overexpression of Gmh1p can compensate in vivo defects caused by mutations in this domain. We show that Gmh1p is an integral membrane protein that localizes to the early Golgi in yeast and in human HeLa cells and cycles through the ER. Hence, we propose that Gmh1p acts as a positive Golgi-membrane partner for Gea function. These results are of general interest given the evolutionary conservation of both ARF-GEFs and the Gmh proteins.
Figures




References
-
- Antony, C., Cibert, C., Geraud, G., Santa Maria, A., Maro, B., Mayau, V., and Goud, B. (1992). The small GTP-binding protein rab6p is distributed from medial Golgi to the trans-Golgi network as determined by a confocal microscopic approach. J. Cell Sci. 103, 785-796. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

