Sequential protein association with nascent 60S ribosomal particles
- PMID: 12808088
- PMCID: PMC164837
- DOI: 10.1128/MCB.23.13.4449-4460.2003
Sequential protein association with nascent 60S ribosomal particles
Abstract
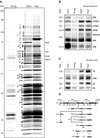
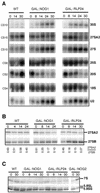
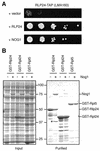
Ribosome biogenesis in eukaryotes depends on the coordinated action of ribosomal and nonribosomal proteins that guide the assembly of preribosomal particles. These intermediate particles follow a maturation pathway in which important changes in their protein composition occur. The mechanisms involved in the coordinated assembly of the ribosomal particles are poorly understood. We show here that the association of preribosomal factors with pre-60S complexes depends on the presence of earlier factors, a phenomenon essential for ribosome biogenesis. The analysis of the composition of purified preribosomal complexes blocked in maturation at specific steps allowed us to propose a model of sequential protein association with, and dissociation from, early pre-60S complexes for several preribosomal factors such as Mak11, Ssf1, Rlp24, Nog1, and Nog2. The presence of either Ssf1 or Nog2 in complexes that contain the 27SB pre-rRNA defines novel, distinct pre-60S particles that contain the same pre-rRNA intermediates and that differ only by the presence or absence of specific proteins. Physical and functional interactions between Rlp24 and Nog1 revealed that the assembly steps are, at least in part, mediated by direct protein-protein interactions.
Figures




References
-
- Bachellerie, J. P., B. Michot, M. Nicoloso, A. Balakin, J. Ni, and M. J. Fournier. 1995. Antisense snoRNAs: a family of nucleolar RNAs with long complementarities to rRNA. Trends Biochem. Sci. 20:261-264. - PubMed
-
- Ban, N., P. Nissen, J. Hansen, P. B. Moore, and T. A. Steitz. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289:905-920. - PubMed
-
- Bassler, J., P. Grandi, O. Gadal, T. Lessmann, E. Petfalski, D. Tollervey, J. Lechner, and E. Hurt. 2001. Identification of a 60S preribosomal particle that is closely linked to nuclear export. Mol. Cell 8:517-529. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
