Targeted cytosine methylation for in vivo detection of protein-DNA interactions
- PMID: 12808133
- PMCID: PMC164658
- DOI: 10.1073/pnas.1332672100
Targeted cytosine methylation for in vivo detection of protein-DNA interactions
Abstract
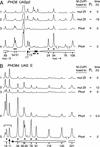
We report a technique, named targeted gene methylation (TAGM), for identifying in vivo protein-binding sites in chromatin. M.CviPI, a cytosine-5 DNA methyltransferase recognizing GC sites, is fused to a DNA-binding factor enabling simultaneous detection of targeted methylation, factor footprints, and chromatin structural changes by bisulfite genomic sequencing. Using TAGM with the yeast transactivator Pho4, methylation enrichments of up to 34- fold occur proximal to native Pho4-binding sites. Additionally, significant selective targeting of methylation is observed several hundred nucleotides away, suggesting the detection of long-range interactions due to higher-order chromatin structure. In contrast, at an extragenic locus lacking Pho4-binding sites, methylation levels are at the detection limit at early times after Pho4 transactivation. Notably, substantial amounts of methylation are targeted by Pho4-M.CviPI under repressive conditions when most of the transactivator is excluded from the nucleus. Thus, TAGM enables rapid detection of DNA-protein interactions even at low occupancies and has potential for identifying factor targets at the genome-wide level. Extension of TAGM from yeast to vertebrates, which use methylation to initiate and propagate repressed chromatin, could also provide a valuable strategy for heritable inactivation of gene expression.
Figures



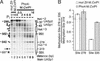
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

