From latent disseminated cells to overt metastasis: genetic analysis of systemic breast cancer progression
- PMID: 12808139
- PMCID: PMC164657
- DOI: 10.1073/pnas.1331931100
From latent disseminated cells to overt metastasis: genetic analysis of systemic breast cancer progression
Abstract
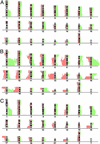
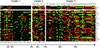
According to the present view, metastasis marks the end in a sequence of genomic changes underlying the progression of an epithelial cell to a lethal cancer. Here, we aimed to find out at what stage of tumor development transformed cells leave the primary tumor and whether a defined genotype corresponds to metastatic disease. To this end, we isolated single disseminated cancer cells from bone marrow of breast cancer patients and performed single-cell comparative genomic hybridization. We analyzed disseminated tumor cells from patients after curative resection of the primary tumor (stage M0), as presumptive progenitors of manifest metastasis, and from patients with manifest metastasis (stage M1). Their genomic data were compared with those from microdissected areas of matched primary tumors. Disseminated cells from M0-stage patients displayed significantly fewer chromosomal aberrations than primary tumors or cells from M1-stage patients (P < 0.008 and P < 0.0001, respectively), and their aberrations appeared to be randomly generated. In contrast, primary tumors and M1 cells harbored different and characteristic chromosomal imbalances. Moreover, applying machine-learning methods for the classification of the genotypes, we could correctly identify the presence or absence of metastatic disease in a patient on the basis of a single-cell genome. We suggest that in breast cancer, tumor cells may disseminate in a far less progressed genomic state than previously thought, and that they acquire genomic aberrations typical of metastatic cells thereafter. Thus, our data challenge the widely held view that the precursors of metastasis are derived from the most advanced clone within the primary tumor.
Figures

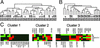


References
-
- Fearon, E. R. & Vogelstein, B. (1990) Cell 61, 759-767. - PubMed
-
- van't Veer, L. J., Dai, H., van de Vijver, M. J., He, Y. D., Hart, A. A., Mao, M., Peterse, H. L., van der Kooy, K., Marton, M. J., Witteveen, A. T., et al. (2002) Nature 415, 530-536. - PubMed
-
- Bernards, R. & Weinberg, R. A. (2002) Nature 418, 823. - PubMed
-
- Hanahan, D. & Weinberg, R. A. (2000) Cell 100, 57-70. - PubMed
-
- Abbruzzese, J. L., Abbruzzese, M. C., Hess, K. R., Raber, M. N., Lenzi, R. & Frost, P. (1994) J. Clin. Oncol. 12, 1272-1280. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

