Crystal structure of the SOS cell division inhibitor SulA and in complex with FtsZ
- PMID: 12808143
- PMCID: PMC164683
- DOI: 10.1073/pnas.1330742100
Crystal structure of the SOS cell division inhibitor SulA and in complex with FtsZ
Abstract
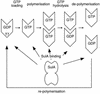
SulA halts cell division in Escherichia coli by binding to the major component of the division machinery FtsZ. We have solved the crystal structure of SulA alone and in complex with FtsZ from Pseudomonas aeruginosa. SulA is expressed when the SOS response is induced. This is a mechanism to inhibit cell division and repair DNA in the event of DNA damage. FtsZ is a tubulin-like protein that forms polymers, with the active-site GTPase split across two monomers. One monomer provides the GTP-binding site and the other, through its T7 loop nucleotide hydrolysis. Our structures show that SulA is a dimer, and that SulA inhibits cell division neither by binding the nucleotide-binding site nor by inducing conformational changes in FtsZ. Instead, SulA binds the T7 loop surface of FtsZ, opposite the nucleotide-binding site, blocking polymer formation. These findings explain why GTP hydrolysis and polymer turnover are required for SulA inhibition.
Figures


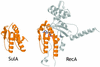
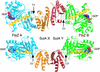

References
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

