The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA
- PMID: 12808465
- PMCID: PMC1350966
- DOI: 10.1038/nature01707
The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA
Abstract
High mutation frequency during reverse transcription has a principal role in the genetic variation of primate lentiviral populations. It is the main driving force for the generation of drug resistance and the escape from immune surveillance. G to A hypermutation is one of the characteristics of primate lentiviruses, as well as other retroviruses, during replication in vivo and in cell culture. The molecular mechanisms of this process, however, remain to be clarified. Here, we demonstrate that CEM15 (also known as apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3G; APOBEC3G), an endogenous inhibitor of human immunodeficiency virus type 1 (HIV-1) replication, is a cytidine deaminase and is able to induce G to A hypermutation in newly synthesized viral DNA. This effect can be counteracted by the HIV-1 virion infectivity factor (Vif). It seems that this viral DNA mutator is a viral defence mechanism in host cells that may induce either lethal hypermutation or instability of the incoming nascent viral reverse transcripts, which could account for the Vif-defective phenotype. Importantly, the accumulation of CEM15-mediated non-lethal hypermutation in the replicating viral genome could potently contribute to the genetic variation of primate lentiviral populations.
Conflict of interest statement
Figures


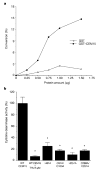
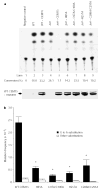
Comment in
-
Good to CU.Nature. 2003 Jul 3;424(6944):21-2. doi: 10.1038/424021a. Nature. 2003. PMID: 12840737 No abstract available.
References
-
- Fitzgibbon JE, Mazar S, Dubin DT. A new type of G → A hypermutation affecting human immunodeficiency virus. AIDS Res Hum Retroviruses. 1993;9:833–838. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

