Characterisation of methionine adenosyltransferase from Mycobacterium smegmatis and M. tuberculosis
- PMID: 12809568
- PMCID: PMC165446
- DOI: 10.1186/1471-2180-3-12
Characterisation of methionine adenosyltransferase from Mycobacterium smegmatis and M. tuberculosis
Abstract
Background: Tuberculosis remains a serious world-wide health threat which requires the characterisation of novel drug targets for the development of future antimycobacterials. One of the key obstacles in the definition of new targets is the large variety of metabolic alterations that occur between cells in the active growth and chronic/dormant phases of tuberculosis. The ideal biochemical target should be active in both growth phases. Methionine adenosyltransferase, which catalyses the formation of S-adenosylmethionine from methionine and ATP, is involved in polyamine biosynthesis during active growth and is also required for the methylation and cyclopropylation of mycolipids necessary for survival in the chronic phase.
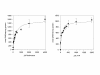
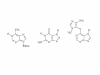
Results: The gene encoding methionine adenosyltransferase has been cloned from Mycobacterium tuberculosis and the model organism M. smegmatis. Both enzymes retained all amino acids known to be involved in catalysing the reaction. While the M. smegmatis enzyme could be functionally expressed, the M. tuberculosis homologue was insoluble and inactive under a large variety of expression conditions. For the M. smegmatis enzyme, the Vmax for S-adenosylmethionine formation was 1.30 micromol/min/mg protein and the Km for methionine and ATP was 288 microM and 76 microM respectively. In addition, the enzyme was competitively inhibited by 8-azaguanine and azathioprine with a Ki of 4.7 mM and 3.7 mM respectively. Azathioprine inhibited the in vitro growth of M. smegmatis with a minimal inhibitory concentration (MIC) of 500 microM, while the MIC for 8-azaguanine was >1.0 mM.
Conclusion: The methionine adenosyltransferase from both organisms had a primary structure very similar those previously characterised in other prokaryotic and eukaryotic organisms. The kinetic properties of the M. smegmatis enzyme were also similar to known prokaryotic methionine adenosyltransferases. Inhibition of the enzyme by 8-azaguanine and azathioprine provides a starting point for the synthesis of higher affinity purine-based inhibitors.
Figures



References
-
- Besra GS, Chatterjee D. Lipids and carbohydrates of Mycobacterium tuberculosis. In: Bloom BR, editor. Tuberculosis. Pathogenesis, protection, and control. Washington, ASM Press; 1994. pp. 285–306.
-
- McKinney JD, Honer zu Bentrup K, Munoz-Elias EJ, Miczak A, Chen B, Chan WT, Swenson D, Sacchettini JC, Jacobs WR, Jr, Russell DG. Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature. 2000;406:735–8. doi: 10.1038/35021074. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

