A snoRNA that guides the two most conserved pseudouridine modifications within rRNA confers a growth advantage in yeast
- PMID: 12810910
- PMCID: PMC1370443
- DOI: 10.1261/rna.5240503
A snoRNA that guides the two most conserved pseudouridine modifications within rRNA confers a growth advantage in yeast
Abstract
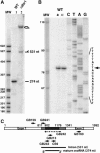
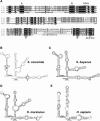
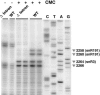
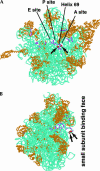
Ribosomal RNAs contain a number of modified nucleotides. The most abundant nucleotide modifications found within rRNAs fall into two types: 2'-O-ribose methylations and pseudouridylations. In eukaryotes, small nucleolar guide RNAs, the snoRNAs that are the RNA components of the snoRNPs, specify the position of these modifications. The 2'-O-ribose methylations and pseudouridylations are guided by the box C/D and box H/ACA snoRNAs, respectively. The role of these modifications in rRNA remains poorly understood as no clear phenotype has yet been assigned to the absence of specific 2'-O-ribose methylations or pseudouridylations. Only very recently, a slight translation defect and perturbation of polysome profiles was reported in yeast for the absence of the Psi at position 2919 within the LSU rRNA. Here we report the identification and characterization in yeast of a novel intronic H/ACA snoRNA that we called snR191 and that guides pseudouridylation at positions 2258 and 2260 in the LSU rRNA. Most interestingly, these two modified bases are the most conserved pseudouridines from bacteria to human in rRNA. The corresponding human snoRNA is hU19. We show here that, in yeast, the presence of this snoRNA, and hence, most likely, of the conserved pseudouridines it specifies, is not essential for viability but provides a growth advantage to the cell.
Figures

Similar articles
-
The complete set of H/ACA snoRNAs that guide rRNA pseudouridylations in Saccharomyces cerevisiae.RNA. 2005 Jun;11(6):928-38. doi: 10.1261/rna.2100905. RNA. 2005. PMID: 15923376 Free PMC article.
-
Different mechanisms for pseudouridine formation in yeast 5S and 5.8S rRNAs.Mol Cell Biol. 2008 May;28(10):3089-100. doi: 10.1128/MCB.01574-07. Epub 2008 Mar 10. Mol Cell Biol. 2008. PMID: 18332121 Free PMC article.
-
Genome-wide searching for pseudouridylation guide snoRNAs: analysis of the Saccharomyces cerevisiae genome.Nucleic Acids Res. 2004 Aug 11;32(14):4281-96. doi: 10.1093/nar/gkh768. Print 2004. Nucleic Acids Res. 2004. PMID: 15306656 Free PMC article.
-
Structure and biogenesis of small nucleolar RNAs acting as guides for ribosomal RNA modification.Acta Biochim Pol. 1999;46(2):377-89. Acta Biochim Pol. 1999. PMID: 10547039 Review.
-
The expanding snoRNA world.Biochimie. 2002 Aug;84(8):775-90. doi: 10.1016/s0300-9084(02)01402-5. Biochimie. 2002. PMID: 12457565 Review.
Cited by
-
Identification of determinants in the protein partners aCBF5 and aNOP10 necessary for the tRNA:Psi55-synthase and RNA-guided RNA:Psi-synthase activities.Nucleic Acids Res. 2007;35(16):5610-24. doi: 10.1093/nar/gkm606. Epub 2007 Aug 17. Nucleic Acids Res. 2007. PMID: 17704128 Free PMC article.
-
Structural and evolutionary insights into ribosomal RNA methylation.Nat Chem Biol. 2018 Feb 14;14(3):226-235. doi: 10.1038/nchembio.2569. Nat Chem Biol. 2018. PMID: 29443970 Review.
-
Nucleotide modifications in three functionally important regions of the Saccharomyces cerevisiae ribosome affect translation accuracy.Nucleic Acids Res. 2009 Dec;37(22):7665-77. doi: 10.1093/nar/gkp816. Nucleic Acids Res. 2009. PMID: 19820108 Free PMC article.
-
Functional importance of individual rRNA 2'-O-ribose methylations revealed by high-resolution phenotyping.RNA. 2008 Apr;14(4):649-56. doi: 10.1261/rna.845808. Epub 2008 Feb 6. RNA. 2008. PMID: 18256246 Free PMC article.
-
28S rRNA is inducibly pseudouridylated by the mTOR pathway translational control in CHO cell cultures.J Biotechnol. 2014 Mar 20;174:16-21. doi: 10.1016/j.jbiotec.2014.01.024. Epub 2014 Jan 27. J Biotechnol. 2014. PMID: 24480570 Free PMC article.
References
-
- Bakin, A. and Ofengand, J. 1993. Four newly located pseudouridylate residues in Escherichia coli 23S ribosomal RNA are all at the peptidyltransferase center: Analysis by the application of a new sequencing technique. Biochemistry 32: 9754–9762. - PubMed
-
- Balakin, A.G., Smith, L., and Fournier, M.J. 1996. The RNA world of the nucleolus: Two major families of small RNAs defined by different box elements with related functions. Cell 86: 823–834. - PubMed
-
- Cherry, J.M., Ball, C., Dolinski, K., Dwight, S., Harris, M., Matese, J.C., Sherlock, G., Binkley, G., Jin, H., Weng, S., et al. 2002. Saccharomyces Genome Database. http://genome-www.stanford.edu/Saccharomyces/. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
