TMC and EVER genes belong to a larger novel family, the TMC gene family encoding transmembrane proteins
- PMID: 12812529
- PMCID: PMC165604
- DOI: 10.1186/1471-2164-4-24
TMC and EVER genes belong to a larger novel family, the TMC gene family encoding transmembrane proteins
Abstract
Background: Mutations in the transmembrane cochlear expressed gene 1 (TMC1) cause deafness in human and mouse. Mutations in two homologous genes, EVER1 and EVER2 increase the susceptibility to infection with certain human papillomaviruses resulting in high risk of skin carcinoma. Here we report that TMC1, EVER1 and EVER2 (now TMC6 and TMC8) belong to a larger novel gene family, which is named TMC for trans membrane channel-like gene family.
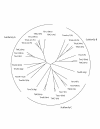
Results: Using a combination of iterative database searches and reverse transcriptase-polymerase chain reaction (RT-PCR) experiments we assembled contigs for cDNA encoding human, murine, puffer fish, and invertebrate TMC proteins. TMC proteins of individual species can be grouped into three subfamilies A, B, and C. Vertebrates have eight TMC genes. The majority of murine TMC transcripts are expressed in most organs; some transcripts, however, in particular the three subfamily A members are rare and more restrictively expressed.
Conclusion: The eight vertebrate TMC genes are evolutionary conserved and encode proteins that form three subfamilies. Invertebrate TMC proteins can also be categorized into these three subfamilies. All TMC genes encode transmembrane proteins with intracellular amino- and carboxyl-termini and at least eight membrane-spanning domains. We speculate that the TMC proteins constitute a novel group of ion channels, transporters, or modifiers of such.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

