Dissection of experimental asthma with DNA microarray analysis identifies arginase in asthma pathogenesis
- PMID: 12813022
- PMCID: PMC161427
- DOI: 10.1172/JCI17912
Dissection of experimental asthma with DNA microarray analysis identifies arginase in asthma pathogenesis
Abstract
Asthma is on the rise despite intense, ongoing research underscoring the need for new scientific inquiry. In an effort to provide unbiased insight into disease pathogenesis, we took an approach involving expression profiling of lung tissue from mice with experimental asthma. Employing asthma models induced by different allergens and protocols, we identified 6.5% of the tested genome whose expression was altered in an asthmatic lung. Notably, two phenotypically similar models of experimental asthma were shown to have distinct transcript profiles. Genes related to metabolism of basic amino acids, specifically the cationic amino acid transporter 2, arginase I, and arginase II, were particularly prominent among the asthma signature genes. In situ hybridization demonstrated marked staining of arginase I, predominantly in submucosal inflammatory lesions. Arginase activity was increased in allergen-challenged lungs, as demonstrated by increased enzyme activity, and increased levels of putrescine, a downstream product. Lung arginase activity and mRNA expression were strongly induced by IL-4 and IL-13, and were differentially dependent on signal transducer and activator of transcription 6. Analysis of patients with asthma supported the importance of this pathway in human disease. Based on the ability of arginase to regulate generation of NO, polyamines, and collagen, these results provide a basis for pharmacologically targeting arginine metabolism in allergic disorders.
Figures

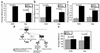


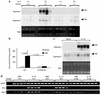

Comment in
-
Arginase: marker, effector, or candidate gene for asthma?J Clin Invest. 2003 Jun;111(12):1815-7. doi: 10.1172/JCI18908. J Clin Invest. 2003. PMID: 12813015 Free PMC article. Review.
References
-
- Holgate ST. The epidemic of allergy and asthma. Nature. 1999;402:B2–B4. - PubMed
-
- Umetsu DT, McIntire JJ, Akbari O, Macaubas C, DeKruyff RH. Asthma: an epidemic of dysregulated immunity. Nat. Immunol. 2002;3:715–720. - PubMed
-
- Broide DH. Molecular and cellular mechanisms of allergic disease. J. Allergy Clin. Immunol. 2001;108:S65–S71. - PubMed
-
- Busse WW, Lemanske RF., Jr Asthma. N. Engl. J. Med. 2001;344:350–362. - PubMed
-
- Lee NA, Gelfand EW, Lee JJ. Pulmonary T cells and eosinophils: coconspirators or independent triggers of allergic respiratory pathology? J. Allergy Clin. Immunol. 2001;107:945–957. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

