Common and distinguishing regulatory and expression characteristics of the highly related KorB proteins of streptomycete plasmids pIJ101 and pSB24.2
- PMID: 12813071
- PMCID: PMC161596
- DOI: 10.1128/JB.185.13.3780-3787.2003
Common and distinguishing regulatory and expression characteristics of the highly related KorB proteins of streptomycete plasmids pIJ101 and pSB24.2
Abstract
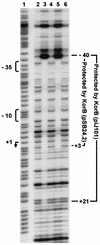
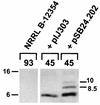
The conjugative plasmid pIJ101 of the spore-forming bacterium Streptomyces lividans contains a regulatory gene, korB, whose product is required to repress potentially lethal expression of the pIJ101 kilB gene. The KorB protein also autoregulates korB gene expression and may be involved in control of pIJ101 copy number. KorB (pIJ101) is expressed as a 10-kDa protein in S. lividans that is immediately processed to a mature 6-kDa repressor molecule. The conjugative Streptomyces cyanogenus plasmid pSB24.1 is deleted upon entry into S. lividans to form pSB24.2, a nonconjugative derivative that contains a korB gene nearly identical to that of pIJ101. Previous evidence that korB of pSB24.2 is capable of overriding pIJ101 kilB-associated lethality supported the notion that pIJ101 and pSB24.2 encode highly related, perhaps even identical conjugation systems. Here we show that KorB (pIJ101) and KorB (pSB24.2) repress transcription from the pIJ101 kilB promoter equally well, although differences exist with respect to their interactions with kilB promoter sequences. Despite high sequence and functional similarities, KorB (pSB24.2) was found to exist as multiple stable forms ranging in size from 10 to 6 kDa both in S. lividans and S. cyanogenus. Immediate processing of KorB (pIJ101) exclusively to the 6-kDa repressor form meanwhile was conserved between the two species. A feature common to both proteins was a marked increase in expression or accumulation upon sporulation, an occurrence that may indicate a particular need for increased quantities of this regulatory protein upon spore germination and resumption of active growth of plasmid-containing cells.
Figures




Similar articles
-
Complementation of conjugation functions of Streptomyces lividans plasmid pIJ101 by the related Streptomyces plasmid pSB24.2.J Bacteriol. 1999 Aug;181(15):4680-5. doi: 10.1128/JB.181.15.4680-4685.1999. J Bacteriol. 1999. PMID: 10419972 Free PMC article.
-
The active form of the KorB protein encoded by the Streptomyces plasmid pIJ101 is a processed product that binds differentially to the two promoters it regulates.J Bacteriol. 1993 Nov;175(21):6996-7005. doi: 10.1128/jb.175.21.6996-7005.1993. J Bacteriol. 1993. PMID: 8226643 Free PMC article.
-
Mutational and functional analysis of the korA and korB gene products of Streptomyces plasmid pIJ101.Mol Gen Genet. 1990 Jul;222(2-3):337-44. doi: 10.1007/BF00633838. Mol Gen Genet. 1990. PMID: 2274034
-
Expression characteristics of the transfer-related kilB gene product of Streptomyces plasmid pIJ101: implications for the plasmid spread function.J Bacteriol. 2001 Feb;183(4):1339-45. doi: 10.1128/JB.183.4.1339-1345.2001. J Bacteriol. 2001. PMID: 11157947 Free PMC article.
-
Conjugative DNA-transfer in Streptomyces, a mycelial organism.Plasmid. 2016 Sep-Nov;87-88:1-9. doi: 10.1016/j.plasmid.2016.09.004. Epub 2016 Sep 28. Plasmid. 2016. PMID: 27687731 Review.
References
-
- Ausubel, F. M., R. Brent, R. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1995. Short protocols in molecular biology, 3rd ed. John Wiley & Sons, New York, N.Y.
-
- Bolotin, A. P., A. V. Sorokin, N. N. Aleksandrov, V. N. Danilenko, and Y. I. Kozlov. 1986. Nucleotide sequence of DNA of the actinomycete plasmid pSB24.2. Dokl. Biochem. 283:260-263. - PubMed
-
- Champness, W. C., and K. F. Chater. 1994. Regulation and integration of antibiotic production and morphological differentiation in Streptomyces spp., p. 61-93. In P. Piggot, J. C. P. Moran, and P. Youngman (ed.), Regulation of bacterial differentiation. American Society for Microbiology, Washington, D.C.
-
- Chater, K. F. 1993. Genetics of differentiation in Streptomyces. Annu. Rev. Microbiol. 47:685-713. - PubMed
-
- Deng, Z., T. Kieser, and D. A. Hopwood. 1988. “Strong incompatibility” between derivatives of the Streptomyces multi-copy plasmid pIJ101. Mol. Gen. Genet. 214:286-294. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

