Molecular analysis of the copper-transporting efflux system CusCFBA of Escherichia coli
- PMID: 12813074
- PMCID: PMC161567
- DOI: 10.1128/JB.185.13.3804-3812.2003
Molecular analysis of the copper-transporting efflux system CusCFBA of Escherichia coli
Abstract
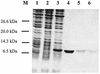
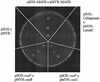
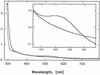
The cus determinant of Escherichia coli encodes the CusCFBA proteins that mediate resistance to copper and silver by cation efflux. CusA and CusB were essential for copper resistance, and CusC and CusF were required for full resistance. Replacements of methionine residues 573, 623, and 672 with isoleucine in CusA resulted in loss of copper resistance, demonstrating their functional importance. Substitutions for several other methionine residues of this protein did not have any effect. The small 10-kDa protein CusF (previously YlcC) was shown to be a periplasmic protein. CusF bound one copper per polypeptide. The pink CusF copper protein complex exhibited an absorption maximum at around 510 nm. Methionine residues of CusF were involved in copper binding as shown by site-directed mutagenesis. CusF interacted with CusB and CusC polypeptides in a yeast two-hybrid assay. In contrast to other well-studied CBA-type heavy metal efflux systems, Cus was shown to be a tetrapartite resistance system that involves the novel periplasmic copper-binding protein CusF. These data provide additional evidence for the hypothesis that Cu(I) is directly transported from the periplasm across the outer membrane by the Cus complex.
Figures

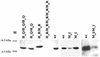
References
-
- Andersen, C., C. Hughes, and V. Koronakis. 2001. Protein export and drug efflux through bacterial channel-tunnels. Curr. Opin. Cell Biol. 13:412-416. - PubMed
-
- Andersen, C., E. Koronakis, C. Hughes, and V. Koronakis. 2002. An aspartate ring at the TolC tunnel entrance determines ion selectivity and presents a target for blocking by large cations. Mol. Microbiol. 44:1131-1139. - PubMed
-
- Anraku, Y., and R. B. Gennis. 1987. The aerobic respiratory chain of Escherichia coli. Trends Biochem. Sci. 12:262-266.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

