Two newly characterized germinal center B-cell-associated genes, GCET1 and GCET2, have differential expression in normal and neoplastic B cells
- PMID: 12819018
- PMCID: PMC1868163
- DOI: 10.1016/S0002-9440(10)63637-1
Two newly characterized germinal center B-cell-associated genes, GCET1 and GCET2, have differential expression in normal and neoplastic B cells
Abstract
A group of genes are highly expressed in normal germinal center (GC) B cells and GC B-cell-derived malignancies based on cDNA microarray analysis. Two new genes, GCET1 (germinal center B-cell expressed transcript 1) and GCET2, were cloned from selected expressed sequence tags (IMAGE clone 1334260 and 814622, respectively). GCET1 is located on chromosome 14q32 and has four splicing isoforms, of which the longest one is 1787 bp and encodes a 435-amino acid protein. GCET2 is located on 3q13.13, and the cloned fragment is 3270 bp, which encodes a protein of 178 amino acids. Blast search showed that GCET1 has a highly conserved serine proteinase inhibitor (SERPIN) domain and is located on a chromosomal locus containing seven other SERPIN family members. GCET2 is a likely homologue of the mouse gene M17, a GC-expressed transcript. Analysis of the GCET2 protein sequence indicated that it may be involved in signal transduction in the cytoplasm. Northern blot and real-time polymerase chain reaction analyses confirmed that GCET1 is highly restricted to normal GC B cells and GCB-cell-derived cell lines. Although GCET2 is also a useful marker for normal and neoplastic GC B cells, it has a wider range of expression including immature B and T cells. Real-time polymerase chain reaction assay showed that both GCET1 and GCET2 are preferentially expressed in follicular lymphoma and diffuse large B-cell lymphoma with GC B-cell differentiation, confirming previous microarray gene expression analysis, but neither one is entirely specific. Multiple markers are necessary to differentiate the GCB from the activated B-cell type of diffuse large B-cell lymphoma with a high degree of accuracy.
Figures
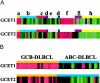





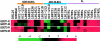
References
-
- Ahmed R, Gray D: Immunological memory and protective immunity: understanding their relation. Science 1996, 272:54-60 - PubMed
-
- Cariappa A, Pillai S: Antigen-dependent B-cell development. Curr Opin Immunol 2002, 14:241-249 - PubMed
-
- Steinman RM: Dendritic cells and the control of immunity: enhancing the efficiency of antigen presentation. Mt Sinai J Med 2001, 68:106-166 - PubMed
-
- Han S, Zheng B, Takahashi Y, Kelsoe G: Distinctive characteristics of germinal center B cells. Semin Immunol 1997, 9:255-260 - PubMed
-
- Kelsoe G: Life and death in germinal centers (redux). Immunity 1996, 4:107-111 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

