Identification of novel cellular targets in biliary tract cancers using global gene expression technology
- PMID: 12819026
- PMCID: PMC1868162
- DOI: 10.1016/S0002-9440(10)63645-0
Identification of novel cellular targets in biliary tract cancers using global gene expression technology
Erratum in
-
Corrections.Am J Pathol. 2017 Apr;187(4):936. doi: 10.1016/j.ajpath.2017.02.002. Am J Pathol. 2017. PMID: 28342446 Free PMC article. No abstract available.
Abstract
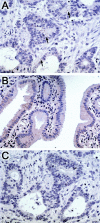
Biliary tract carcinoma carries a poor prognosis, and difficulties with clinical management in patients with advanced disease are often due to frequent late-stage diagnosis, lack of serum markers, and limited information regarding biliary tumor pathogenesis. RNA-based global analyses of gene expression have led to the identification of a large number of up-regulated genes in several cancer types. We have used the recently developed Affymetrix U133A gene expression microarrays containing nearly 22,000 unique transcripts to obtain global gene expression profiles from normal biliary epithelial scrapings (n = 5), surgically resected biliary carcinomas (n = 11), and biliary cancer cell lines (n = 9). Microarray hybridization data were normalized using dCHIP (http://www.dCHIP.org) to identify differentially up-regulated genes in primary biliary cancers and biliary cancer cell lines and their expression profiles was compared to that of normal epithelial scrapings using the dCHIP software as well as Significance Analysis of Microarrays or SAM (http://www-stat.stanford.edu/ approximately tibs/SAM/). Comparison of the dCHIP and SAM datasets revealed an overlapping list of 282 genes expressed at greater than threefold levels in the cancers compared to normal epithelium (t-test P <0.1 in dCHIP, and median false discovery rate <10 in SAM). Several pathways integral to tumorigenesis were up-regulated in the biliary cancers, including proliferation and cell cycle antigens (eg, cyclins D2 and E2, cdc2/p34, and geminin), transcription factors (eg, homeobox B7 and islet-1), growth factors and growth factor receptors (eg, hepatocyte growth factor, amphiregulin, and insulin-like growth factor 1 receptor), and enzymes modulating sensitivity to chemotherapeutic agents (eg, cystathionine beta synthase, dCMP deaminase, and CTP synthase). In addition, we identified several "pathway" genes that are rapidly emerging as novel therapeutic targets in cancer (eg, cytosolic phospholipase A2, an upstream target of the cyclooxygenase pathway, and ribosomal protein S6 kinase and eukaryotic translation initiation factor 4E, two important downstream mediators of the mitogenic Akt/mTOR signaling pathway). Overexpression of selected up-regulated genes was confirmed in tissue microarrays of biliary cancers by immunohistochemical analysis (n = 4) or in situ hybridization (n = 1), and in biliary cancer cell lines by reverse transcriptase PCR (n = 2). The majority of genes identified in the present study has not been previously reported in biliary cancers, and represent novel potential screening and therapeutic targets of this cancer type.
Figures



References
-
- Greenlee RT, Hill-Harmon MB, Murray T, Thun M: Cancer statistics, 2001. CA Cancer J Clin 2001, 51:15-36 - PubMed
-
- de Groen PC, Gores GJ, LaRusso NF, Gunderson LL, Nagorney DM: Biliary tract cancers. N Engl J Med 1999, 341:1368-1378 - PubMed
-
- Lazcano-Ponce EC, Miquel JF, Munoz N, Herrero R, Ferrecio C, Wistuba II, Alonso de Ruiz P, Aristi Urista G, Nervi F: Epidemiology and molecular pathology of gallbladder cancer. CA Cancer J Clin 2001, 51:349-364 - PubMed
-
- Iacobuzio-Donahue CA, Maitra A, Shen-Ong GL, van Heek T, Ashfaq R, Meyer R, Walter K, Berg K, Hollingsworth MA, Cameron JL, Yeo CJ, Kern SE, Goggins M, Hruban RH: Discovery of novel tumor markers of pancreatic cancer using global gene expression technology. Am J Pathol 2002, 160:1239-1249 - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

