Mutation of sarA in Staphylococcus aureus limits biofilm formation
- PMID: 12819120
- PMCID: PMC161964
- DOI: 10.1128/IAI.71.7.4206-4211.2003
Mutation of sarA in Staphylococcus aureus limits biofilm formation
Abstract
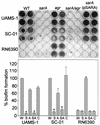
Mutation of sarA resulted in a reduced capacity to form a biofilm in six of the eight Staphylococcus aureus strains we tested (UAMS-1, UAMS-601, SA113, SC-01, S6C, and DB). The exceptions were Newman, which formed a poor biofilm under all conditions, and RN6390, which consistently formed a biofilm only after mutation of agr. Mutation of agr in other strains had little impact on biofilm formation. In every strain other than Newman, including RN6390, simultaneous mutation of sarA and agr resulted in a phenotype like that observed with the sarA mutants. Complementation studies using a sarA clone confirmed that the defect in biofilm formation was due to the sarA mutation.
Figures





References
-
- Arvidson, S., and K. Tegmark. 2001. Regulation of virulence determinants in Staphylococcus aureus. Int. J. Med. Microbiol. 291:159-170. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

