Impact of alternative initiation, splicing, and termination on the diversity of the mRNA transcripts encoded by the mouse transcriptome
- PMID: 12819126
- PMCID: PMC403716
- DOI: 10.1101/gr.1017303
Impact of alternative initiation, splicing, and termination on the diversity of the mRNA transcripts encoded by the mouse transcriptome
Abstract
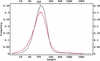
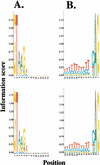
We analyzed the FANTOM2 clone set of 60,770 RIKEN full-length mouse cDNA sequences and 44,122 public mRNA sequences. We developed a new computational procedure to identify and classify the forms of splice variation evident in this data set and organized the results into a publicly accessible database that can be used for future expression array construction, structural genomics, and analyses of the mechanism and regulation of alternative splicing. Statistical analysis shows that at least 41% and possibly as much as 60% of multiexon genes in mouse have multiple splice forms. Of the transcription units with multiple splice forms, 49% contain transcripts in which the apparent use of an alternative transcription start (stop) is accompanied by alternative splicing of the initial (terminal) exon. This implies that alternative transcription may frequently induce alternative splicing. The fact that 73% of all exons with splice variation fall within the annotated coding region indicates that most splice variation is likely to affect the protein form. Finally, we compared the set of constitutive (present in all transcripts) exons with the set of cryptic (present only in some transcripts) exons and found statistically significant differences in their length distributions, the nucleotide distributions around their splice junctions, and the frequencies of occurrence of several short sequence motifs.
Figures

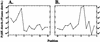

References
-
- Berget, S. 1995. Exon recognition in vertebrate splicing. J. Biol. Chem. 270: 2411-2414. - PubMed
-
- Brett, D., Hanke, J., Lehmann, G., Haase, S., Delbruck, S., Krueger, S., Reich, J., and Bork, P. 2000. EST comparison indicates 38% of human mRNAs contain possible alternative splice forms. FEBS Lett. 474: 83-86. - PubMed
-
- Brett, D., Pospisil, H., Valcarcel, J., Reich, J., and Bork, P. 2002. Alternative splicing and genome complexity. Nat. Genet. 30: 29-30. - PubMed
WEB SITE REFERENCES
-
- ftp://wolfram.wi.mit.edu/pub/mousecontigs/MGSCV3; draft of the mouse genome sequence.
-
- http://facts.gsc.riken.go.jp; Functional Association/annotation of cDNA clones from Text/sequence Sources (FACTS).
-
- http://genomes.rockefeller.edu/MouSDB; database of alternative splice forms in the mouse transcriptome.
-
- http://smart.embl-heidelberg.de; Simple Modular Architecture Research Tool (SMART). - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
