Mouse proteome analysis
- PMID: 12819131
- PMCID: PMC403658
- DOI: 10.1101/gr.978703
Mouse proteome analysis
Abstract
A general overview of the protein sequence set for the mouse transcriptome produced during the FANTOM2 sequencing project is presented here. We applied different algorithms to characterize protein sequences derived from a nonredundant representative protein set (RPS) and a variant protein set (VPS) of the mouse transcriptome. The functional characterization and assignment of Gene Ontology terms was done by analysis of the proteome using InterPro. The Superfamily database analyses gave a detailed structural classification according to SCOP and provide additional evidence for the functional characterization of the proteome data. The MDS database analysis revealed new domains which are not presented in existing protein domain databases. Thus the transcriptome gives us a unique source of data for the detection of new functional groups. The data obtained for the RPS and VPS sets facilitated the comparison of different patterns of protein expression. A comparison of other existing mouse and human protein sequence sets (e.g., the International Protein Index) demonstrates the common patterns in mammalian proteomes. The analysis of the membrane organization within the transcriptome of multiple eukaryotes provides valuable statistics about the distribution of secretory and transmembrane proteins
Figures
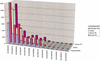

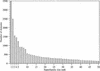

References
-
- Apweiler, R., Attwood, T.K., Bairoch, A., Bateman, A., Birney, E., Biswas, M., Bucher, P., Cerutti, L., Corpet, F., Croning, M.D., et al. 2001. The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res. 29: 37-40. - PMC - PubMed
-
- FANTOM Consortium 2002. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature 420: 563-573. - PubMed
-
- Gough, J. 2002. The superfamily database in structural genomics. Acta Crystallogr. D Biol. Crystallogr. 58: 1897-1900. - PubMed
WEB SITE REFERENCES
-
- http://www.celeradiscoverysystem.com; Celera Corporation.
-
- http://www.ensembl.org/; ENSEMBL genome databases.
-
- http://www.ebi.ac.uk/IPI; International Protein Index.
-
- http://www.ebi.ac.uk/proteome; Proteome Analysis Database.
-
- http://genome.gsc.riken.go.jp/; RIKEN Mouse Representative Transcript and Protein Sets. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
