Exploration of the cell-cycle genes found within the RIKEN FANTOM2 data set
- PMID: 12819135
- PMCID: PMC403664
- DOI: 10.1101/gr.1012403
Exploration of the cell-cycle genes found within the RIKEN FANTOM2 data set
Abstract
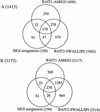
The cell cycle is one of the most fundamental processes within a cell. Phase-dependent expression and cell-cycle checkpoints require a high level of control. A large number of genes with varying functions and modes of action are responsible for this biology. In a targeted exploration of the FANTOM2-Variable Protein Set, a number of mouse homologs to known cell-cycle regulators as well as novel members of cell-cycle families were identified. Focusing on two prototype cell-cycle families, the cyclins and the NIMA-related kinases (NEKs), we believe we have identified all of the mouse members of these families, 24 cyclins and 10 NEKs, and mapped them to ENSEMBL transcripts. To attempt to globally identify all potential cell cycle-related genes within mouse, the MGI (Mouse Genome Database) assignments for the RIKEN Representative Set (RPS) and the results from two homology-based queries were merged. We identified 1415 genes with possible cell-cycle roles, and 1758 potential paralogs. We comment on the genes identified in this screen and evaluate the merits of each approach.
Figures

References
-
- Apweiler, R., Attwood, T.K., Bairoch, A., Bateman, A., Birney, E., Biswas, M., Bucher, P., Cerutti, L., Corpet, F., Croning, M.D.R., et al. 2001. The InterPro database, an integrated documentation resource for protein families, domains, and functional sites. Nucleic Acids Res. 29: 37-40. - PMC - PubMed
-
- Berke, J.D., Sgambato, V., Zhu, P.P., Lavoie, B., Vincent, M., Krause, M., and Hyman, S.E. 2001. Dopamine and glutamate induce distinct striatal splice forms of Ania-6, an RNA polymerase II-associated cyclin. Neuron 32: 277-287. - PubMed
-
- Cho, R.J., Campbell, M.J., Winzeler, E.A., Steinmetz, L., Conway, A., Wodicka, L., Wolfsberg, T.G., Gabrielian, A.E., Landsman, D., Lockhart, D.J., et al. 1998. A genome-wide transcriptional analysis of the mitotic cell cycle. Mol. Cell 2: 65-73. - PubMed
WEB SITE REFERENCES
-
- http://www.godatabase.org/cgi-bin/go.cgi; AMIGO gene ontology browser.
-
- http://www.ncbi.nih.gov/BLAST; Basic Local Alignment Search Tool homepage. - PubMed
-
- http://cmap.nci.nih.gov/Ontologies; Cancer Analysis Molecular Project homepage.
-
- http://www.ensembl.org/Mus_musculus; ENSEMBL mouse genome homepage.
-
- http://fantom2.gsc.riken.go.jp; Functional Annotation of Mouse, RIKEN.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
