A comprehensive transcript map of the mouse Gnas imprinted complex
- PMID: 12819140
- PMCID: PMC403675
- DOI: 10.1101/gr.955503
A comprehensive transcript map of the mouse Gnas imprinted complex
Abstract
The recent publication of the FANTOM mouse transcriptome has provided a unique opportunity to study the diversity of transcripts arising from a single gene locus. We have focused on the Gnas complex, as imprinting loci themselves provide unique insights into transcriptional regulation. Thirteen full-length cDNAs from the FANTOM2 set were mapped to the Gnas locus. These represented one previously described transcript and 12 putative new transcripts. Of these, eight were found to be differentially expressed from either the maternal or paternal allele. Two clones extended Nespas in the 3' direction, providing evidence of antisense transcription spanning a 30-kb genomic region from a single allele. The transcripts were summarized into six transcriptional units, Nespas, Nesp, Gnasxl, F7, exon 1A, and Gnas. The resolution of the Gnas transcript map by the FANTOM2 clones revealed a pattern of alternate splicing. In addition to the transcripts described previously as splicing onto exon 2 of Gnas, each new sense transcript had an alternate short 3'UTR independent of Gnas. Both spliced and unspliced variants of the new imprinted sense transcripts were found. Whereas the functional significance of these alternate transcripts is not known, the availability of the FANTOM clones has provided remarkable insights into the repertoire of transcripts in the Gnas complex locus.
Figures
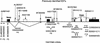


References
-
- Bonthron, D.T., Hayward, B.E., Moran, V., and Strain, L. 2000. Characterization of TH1 and CTSZ, two non-imprinted genes downstream of GNAS1 in chromosome 20q13. Hum. Genet. 107: 165-175. - PubMed
-
- Cattanach, B.M. and Kirk, M. 1985. Differential activity of maternally and paternally derived chromosome regions in mice. Nature 315: 496-498. - PubMed
-
- Cattanach, B.M., Peters, J., Ball, S., and Rasberry, C. 2000. Two imprinted gene mutations: Three phenotypes. Hum. Mol. Genet. 9: 2263-2273. - PubMed
-
- Crawford, J.A., Mutchler, K.J., Sullivan, B.E., Lanigan, T.M., Clark, M.S., and Russo, A.F. 1993. Neural expression of a novel alternatively spliced and polyadenylated Gs α transcript. J. Biol. Chem. 268: 9879-9885. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
