Systematic characterization of the zinc-finger-containing proteins in the mouse transcriptome
- PMID: 12819142
- PMCID: PMC403681
- DOI: 10.1101/gr.949803
Systematic characterization of the zinc-finger-containing proteins in the mouse transcriptome
Abstract
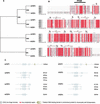
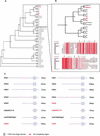
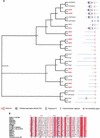
Zinc-finger-containing proteins can be classified into evolutionary and functionally divergent protein families that share one or more domains in which a zinc ion is tetrahedrally coordinated by cysteines and histidines. The zinc finger domain defines one of the largest protein superfamilies in mammalian genomes;46 different conserved zinc finger domains are listed in InterPro (http://www.ebi.ac.uk/InterPro). Zinc finger proteins can bind to DNA, RNA, other proteins, or lipids as a modular domain in combination with other conserved structures. Owing to this combinatorial diversity, different members of zinc finger superfamilies contribute to many distinct cellular processes, including transcriptional regulation, mRNA stability and processing, and protein turnover. Accordingly, mutations of zinc finger genes lead to aberrations in a broad spectrum of biological processes such as development, differentiation, apoptosis, and immunological responses. This study provides the first comprehensive classification of zinc finger proteins in a mammalian transcriptome. Specific detailed analysis of the SP/Krüppel-like factors and the E3 ubiquitin-ligase RING-H2 families illustrates the importance of such an analysis for a more comprehensive functional classification of large protein families. We describe the characterization of a new family of C2H2 zinc-finger-containing proteins and a new conserved domain characteristic of this family, the identification and characterization of Sp8, a new member of the Sp family of transcriptional regulators, and the identification of five new RING-H2 proteins.
Figures
References
-
- Aasland, R., Gibson, T.G., and Stewart, A.F. 1995. The PHD finger: Implications for chromatin-mediated transcriptional regulation. Trends Biochem. Sci. 20: 56-59. - PubMed
-
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403-410. - PubMed
-
- Bach, I. 2000. The LIM domain: Regulation by association. Mech. Dev. 91: 5-17. - PubMed
-
- Bach, I., Rodriguez-Esteban, C., Carriere, C., Bhushan, A., Krones, A., Rose, D.W., Glass, C.K., Andersen, B., Izpisua Belmonte, J.C., and Rosenfeld, M.G. 1999. RLIM inhibits functional activity of LIM homeodomain transcription factors via recruitment of the histone deacetylase complex. Nat. Genet. 22: 394-399. - PubMed
-
- Baker, S.J. and Reddy, E.P. 2000. Cloning of murine G1RP, a novel gene related to Drosophila melanogaster g1. Gene 248: 33-40. - PubMed
WEB SITE REFERENCES
-
- ftp://ftp.ncbi.nih.gov/refseq/; The NCBI Reference Sequence project (RefSeq).
-
- http://cassandra.visac.uq.edu.au/zf; RTPS zinc finger data set.
-
- http://fantom2.gsc.riken.go.jp/; FANTOM 2.
-
- http://genome.gsc.riken.go.jp; Genome Exploration Research Group.
-
- http://genomes.rockefeller.edu; Laboratory of Computational Genomics.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
