The mammalian protein-protein interaction database and its viewing system that is linked to the main FANTOM2 viewer
- PMID: 12819152
- PMCID: PMC403706
- DOI: 10.1101/gr.956303
The mammalian protein-protein interaction database and its viewing system that is linked to the main FANTOM2 viewer
Abstract
Here, we describe the development of a mammalian protein-protein interaction (PPI) database and of a PPI Viewer application to display protein interaction networks (http://fantom21.gsc.riken.go.jp/PPI/). In the database, we stored the mammalian PPIs identified through our PPI assays (internal PPIs), as well as those we extracted and processed (external PPIs) from publicly available data sources, the DIP and BIND databases and MEDLINE abstracts by using FACTS, a new functional inference and curation system. We integrated the internal and external PPIs into the PPI database, which is linked to the main FANTOM2 viewer. In addition, we incorporated into the PPI Viewer information regarding the luciferase reporter activity of internal PPIs and the data confidence of external PPIs; these data enable visualization and evaluation of the reliability of each interaction. Using the described system, we successfully identified several interactions of biological significance. Therefore, the PPI Viewer is a useful tool for exploring FANTOM2 clone-related protein interactions and their potential effects on signaling and cellular communication.
Figures
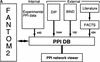
ppi_primers: Contains primer IDs as well as primer sequences and clone IDs. A protein sequence can be estimated by using this table and the RIKEN cDNA database.
ppi_function: Includes the functional annotation for each RIKEN clone.
interaction: Comprises a list of interactions identified through our assays (internal PPIs).
ext_interaction: Stores a list of interactions extracted from publicly available data sources (external PPIs).
ext_protein: Includes protein information of the external PPIs. Each protein is identified by its NCBI accession number.
ppi_source: Stores information regarding the origin of the interactions, i.e., RIKEN_PPI assay, DIP, BIND, or literature.
homology: Contains the results of homology searches and the alignment of two sequences.

ppi_primers: Contains primer IDs as well as primer sequences and clone IDs. A protein sequence can be estimated by using this table and the RIKEN cDNA database.
ppi_function: Includes the functional annotation for each RIKEN clone.
interaction: Comprises a list of interactions identified through our assays (internal PPIs).
ext_interaction: Stores a list of interactions extracted from publicly available data sources (external PPIs).
ext_protein: Includes protein information of the external PPIs. Each protein is identified by its NCBI accession number.
ppi_source: Stores information regarding the origin of the interactions, i.e., RIKEN_PPI assay, DIP, BIND, or literature.
homology: Contains the results of homology searches and the alignment of two sequences.




References
-
- Adams, M.D., Celniker, S.E., Holt, R.A., Evans, C.A., Gocayne, J.D., Amanatides, P.G., Scherer, S.E., Li, P.W., Hoskins, R.A., Galle, R.F., et al. 2000. The genome sequence of Drosophila melanogaster. Science 287: 2185-2195. - PubMed
-
- Antonny, B. and Schekman, R. 2001. ER export: Public transportation by the COPII coach. Curr. Opin. Cell. Biol. 13: 438-443. - PubMed
-
- Castro, P., Liang, H., Liang, J.C., and Nagarajan, L. 2002. A novel, evolutionarily conserved gene family with putative sequence-specific single-stranded DNA-binding activity. Genomics 80: 78-85. - PubMed
-
- The C. elegans Sequencing Consortium. 1998. Genome sequence of the nematode C. elegans: A platform for investigating biology. Science 282: 2012-2018. - PubMed
WEB SITE REFERENCES
-
- http://fantom21.gsc.riken.go.jp/PPI/; PPI Viewer.
-
- http://facts.gsc.riken.go.jp; FACTS system.
-
- http://dip.doe-mbi.ucla.edu; DIP database.
-
- http://www.binddb.org/; BIND database.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
