AGRIS: Arabidopsis gene regulatory information server, an information resource of Arabidopsis cis-regulatory elements and transcription factors
- PMID: 12820902
- PMCID: PMC166152
- DOI: 10.1186/1471-2105-4-25
AGRIS: Arabidopsis gene regulatory information server, an information resource of Arabidopsis cis-regulatory elements and transcription factors
Abstract
Background: The gene regulatory information is hardwired in the promoter regions formed by cis-regulatory elements that bind specific transcription factors (TFs). Hence, establishing the architecture of plant promoters is fundamental to understanding gene expression. The determination of the regulatory circuits controlled by each TF and the identification of the cis-regulatory sequences for all genes have been identified as two of the goals of the Multinational Coordinated Arabidopsis thaliana Functional Genomics Project by the Multinational Arabidopsis Steering Committee (June 2002).
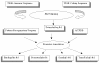
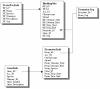
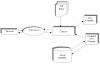
Results: AGRIS is an information resource of Arabidopsis promoter sequences, transcription factors and their target genes. AGRIS currently contains two databases, AtTFDB (Arabidopsis thaliana transcription factor database) and AtcisDB (Arabidopsis thaliana cis-regulatory database). AtTFDB contains information on approximately 1,400 transcription factors identified through motif searches and grouped into 34 families. AtTFDB links the sequence of the transcription factors with available mutants and, when known, with the possible genes they may regulate. AtcisDB consists of the 5' regulatory sequences of all 29,388 annotated genes with a description of the corresponding cis-regulatory elements. Users can search the databases for (i) promoter sequences, (ii) a transcription factor, (iii) a direct target genes for a specific transcription factor, or (vi) a regulatory network that consists of transcription factors and their target genes.
Conclusion: AGRIS provides the necessary software tools on Arabidopsis transcription factors and their putative binding sites on all genes to initiate the identification of transcriptional regulatory networks in the model dicotyledoneous plant Arabidopsis thaliana. AGRIS can be accessed from http://arabidopsis.med.ohio-state.edu.
Figures


References
-
- Reichmann J, Heard J, Martin G, Reuber L, Jiang C-Z, Keddie J, Adam L, Pineada O, Ratcliffe O, Samah R, Creelman R, Pilgrim M, Broun P, Zhang J, Ghandehari D, Sherman B, Yu G-L. Arabidopsis Transcription Factors: Genome-Wide Caomparative Analysis Among Eukaryotes. Science. 2000;209:2105–2110. doi: 10.1126/science.290.5499.2105. - DOI - PubMed
-
- Chen W, Provart N, Glazebrook J, Katagiri F, Chang H, Eulgem T, Mauch F, Luan S, Zou G, Whitham S, Budworth P, Tao Y, Xie Z, Chen X, Lam S, Kreps J, Harper J, Si-Ammour A, Mauch-Mani B, Heinlein M, Kobayashi K, Hohn T, Dangl J, Wang X, Zhu T. Expression profile matrix of Arabidopsis transcription factor genes suggests their putative functions in response to environmental stresses. Plant Cell. 2002;14:559–574. doi: 10.1105/tpc.010410. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

