Reproducibility of gene expression across generations of Affymetrix microarrays
- PMID: 12823866
- PMCID: PMC165600
- DOI: 10.1186/1471-2105-4-27
Reproducibility of gene expression across generations of Affymetrix microarrays
Abstract
Background: The development of large-scale gene expression profiling technologies is rapidly changing the norms of biological investigation. But the rapid pace of change itself presents challenges. Commercial microarrays are regularly modified to incorporate new genes and improved target sequences. Although the ability to compare datasets across generations is crucial for any long-term research project, to date no means to allow such comparisons have been developed. In this study the reproducibility of gene expression levels across two generations of Affymetrix GeneChips (HuGeneFL and HG-U95A) was measured.
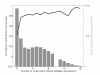
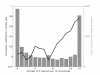
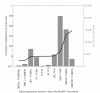
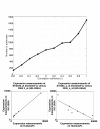
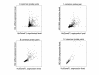
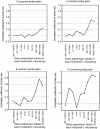
Results: Correlation coefficients were computed for gene expression values across chip generations based on different measures of similarity. Comparing the absolute calls assigned to the individual probe sets across the generations found them to be largely unchanged.
Conclusion: We show that experimental replicates are highly reproducible, but that reproducibility across generations depends on the degree of similarity of the probe sets and the expression level of the corresponding transcript.
Figures
References
-
- Lockhart DJ, Dong H, Byrne MC, Follettie MT, Gallo MV, Chee MS, Mittmann M, Wang C, Kobayashi M, Horton H, et al. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat Biotechnol. 1996;14:1675–80. - PubMed
-
- Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–70. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

