CASTp: Computed Atlas of Surface Topography of proteins
- PMID: 12824325
- PMCID: PMC168919
- DOI: 10.1093/nar/gkg512
CASTp: Computed Atlas of Surface Topography of proteins
Abstract
Computed Atlas of Surface Topography of proteins (CASTp) provides an online resource for locating, delineating and measuring concave surface regions on three-dimensional structures of proteins. These include pockets located on protein surfaces and voids buried in the interior of proteins. The measurement includes the area and volume of pocket or void by solvent accessible surface model (Richards' surface) and by molecular surface model (Connolly's surface), all calculated analytically. CASTp can be used to study surface features and functional regions of proteins. CASTp includes a graphical user interface, flexible interactive visualization, as well as on-the-fly calculation for user uploaded structures. CASTp is updated daily and can be accessed at http://cast.engr.uic.edu.
Figures
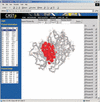
Similar articles
-
CASTp 3.0: computed atlas of surface topography of proteins.Nucleic Acids Res. 2018 Jul 2;46(W1):W363-W367. doi: 10.1093/nar/gky473. Nucleic Acids Res. 2018. PMID: 29860391 Free PMC article.
-
CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues.Nucleic Acids Res. 2006 Jul 1;34(Web Server issue):W116-8. doi: 10.1093/nar/gkl282. Nucleic Acids Res. 2006. PMID: 16844972 Free PMC article.
-
CASTpFold: Computed Atlas of Surface Topography of the universe of protein Folds.Nucleic Acids Res. 2024 Jul 5;52(W1):W194-W199. doi: 10.1093/nar/gkae415. Nucleic Acids Res. 2024. PMID: 38783102 Free PMC article.
-
pvSOAR: detecting similar surface patterns of pocket and void surfaces of amino acid residues on proteins.Nucleic Acids Res. 2004 Jul 1;32(Web Server issue):W555-8. doi: 10.1093/nar/gkh390. Nucleic Acids Res. 2004. PMID: 15215448 Free PMC article.
-
Chapter 4. Predicting and characterizing protein functions through matching geometric and evolutionary patterns of binding surfaces.Adv Protein Chem Struct Biol. 2008;75:107-41. doi: 10.1016/S0065-3233(07)75004-0. Epub 2009 Feb 26. Adv Protein Chem Struct Biol. 2008. PMID: 20731991 Free PMC article. Review.
Cited by
-
Prediction of temperature factors from protein sequence.Bioinformation. 2013;9(3):134-40. doi: 10.6026/97320630009134. Epub 2013 Feb 6. Bioinformation. 2013. PMID: 23422595 Free PMC article.
-
In silico comparative structural and compositional analysis of glycoproteins of RSV to study the nature of stability and transmissibility of RSV A.Syst Microbiol Biomanuf. 2023;3(2):312-327. doi: 10.1007/s43393-022-00110-x. Epub 2022 May 27. Syst Microbiol Biomanuf. 2023. PMID: 38013803 Free PMC article.
-
Bioinsecticidal activity of actinomycete secondary metabolites against the acetylcholinesterase of the legume's insect pest Acyrthosiphon pisum: a computational study.J Genet Eng Biotechnol. 2022 Nov 22;20(1):158. doi: 10.1186/s43141-022-00442-0. J Genet Eng Biotechnol. 2022. PMID: 36417041 Free PMC article.
-
X-ray structures of two proteins belonging to Pfam DUF178 revealed unexpected structural similarity to the DUF191 Pfam family.BMC Struct Biol. 2007 Oct 1;7:62. doi: 10.1186/1472-6807-7-62. BMC Struct Biol. 2007. PMID: 17908300 Free PMC article.
-
A Database of Predicted Binding Sites for Cholesterol on Membrane Proteins, Deep in the Membrane.Biophys J. 2018 Aug 7;115(3):522-532. doi: 10.1016/j.bpj.2018.06.022. Epub 2018 Jun 26. Biophys J. 2018. PMID: 30007584 Free PMC article.
References
-
- Edeslbrunner H., Facello,M. and Liang,J. (1998) On the definition and the construction of pockets in macromolecules. Disc. Appl. Math., 88, 18–29. - PubMed
-
- Lee B. and Richards,F.M. (1971) The interpretation of protein structures: estimation of static accessibility. J. Mol. Biol., 55, 379–400. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

