LGA: A method for finding 3D similarities in protein structures
- PMID: 12824330
- PMCID: PMC168977
- DOI: 10.1093/nar/gkg571
LGA: A method for finding 3D similarities in protein structures
Abstract
We present the LGA (Local-Global Alignment) method, designed to facilitate the comparison of protein structures or fragments of protein structures in sequence dependent and sequence independent modes. The LGA structure alignment program is available as an online service at http://PredictionCenter.llnl.gov/local/lga. Data generated by LGA can be successfully used in a scoring function to rank the level of similarity between two structures and to allow structure classification when many proteins are being analyzed. LGA also allows the clustering of similar fragments of protein structures.
Figures
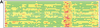



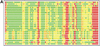

References
-
- Holm L. and Sander,C. (1993) Protein structure comparison by alignment of distance matrices. J. Mol. Biol., 233, 123–138. - PubMed
-
- Feng Z.K. and Sippl,M.J. (1996) Optimum superimposition of protein structures: ambiguities and implications. Fold Des., 1, 123–132. - PubMed
-
- Zemla A., Venclovas,C., Reinhardt,A., Fidelis,K. and Hubbard,T.J. (1997) Numerical criteria for the evaluation of ab initio predictions of protein structure. Proteins, S1, 140–150. - PubMed
-
- Zemla A., Venclovas,C., Moult,J. and Fidelis,K. (1999) Processing and analysis of CASP3 protein structure predictions. Proteins, S3, 22–29. - PubMed
-
- Orengo C.A., Bray,J.E., Hubbard,T., LoConte,L. and Sillitoe,I. (1999) Analysis and assessment of ab initio three-dimensional prediction, secondary structure, and contacts prediction. Proteins, S3, 149–170. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

