GEPAS: A web-based resource for microarray gene expression data analysis
- PMID: 12824345
- PMCID: PMC168997
- DOI: 10.1093/nar/gkg591
GEPAS: A web-based resource for microarray gene expression data analysis
Abstract
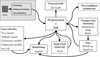
We present a web-based pipeline for microarray gene expression profile analysis, GEPAS, which stands for Gene Expression Profile Analysis Suite (http://gepas.bioinfo.cnio.es). GEPAS is composed of different interconnected modules which include tools for data pre-processing, two-conditions comparison, unsupervised and supervised clustering (which include some of the most popular methods as well as home made algorithms) and several tests for differential gene expression among different classes, continuous variables or survival analysis. A multiple purpose tool for data mining, based on Gene Ontology, is also linked to the tools, which constitutes a very convenient way of analysing clustering results. On-line tutorials are available from our main web server (http://bioinfo.cnio.es).
Figures

References
-
- Lockhart D.J., Dong,H., Byrne,M.C., Follettie,M.T., Gallo,M.V., Chee,M.S., Mittmann,M., Wang,C., Kobayashi,M., Horton,H. and Brown,E.L. (1996) Expression monitoring by hybridisation to high-density oligonucleotide arrays. Nat. Biotechnol., 14, 1675–1680. - PubMed
-
- Stoeckert C.J., Causton,H.C. and Ball,C.A. (2002) Microarray databases: standards and ontologies. Nature Genet., 32, 469–473. - PubMed
-
- Brazma A. and Vilo,J. (2000) Gene expression data analysis. FEBS Lett., 480, 17–24. - PubMed
-
- Herrero J., Díaz-Uriarte,R. and Dopazo,J. (2003) Gene expression data preprocessing. Bioinformatics, 19, 655–656. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

