ChipInfo: Software for extracting gene annotation and gene ontology information for microarray analysis
- PMID: 12824349
- PMCID: PMC169004
- DOI: 10.1093/nar/gkg598
ChipInfo: Software for extracting gene annotation and gene ontology information for microarray analysis
Abstract
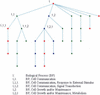
To date, assembling comprehensive annotation information for all probe sets of any Affymetrix microarrays remains a time-consuming, error-prone and challenging task. ChipInfo is designed for retrieving annotation information from online databases such as NetAffx and Gene Ontology and organizing such information into easily interpretable tabular format outputs. As companion software to dChip and GoSurfer, ChipInfo enables users to independently update the information resource files of these software packages. It also has functions for computing related summary statistics of probe sets and Gene Ontology terms. ChipInfo is available at http://biosun1.harvard.edu/complab/chipinfo/.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

