MAK, a computational tool kit for automated MITE analysis
- PMID: 12824388
- PMCID: PMC168938
- DOI: 10.1093/nar/gkg531
MAK, a computational tool kit for automated MITE analysis
Abstract
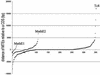
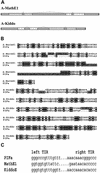
Miniature inverted repeat transposable elements (MITEs) are ubiquitous and numerous in higher eukaryotic genomes. Analysis of MITE families is laborious and time consuming, especially when multiple MITE families are involved in the study. Based on the structural characteristics of MITEs and genetic principles for transposable elements (TEs), we have developed a computational tool kit named MITE analysis kit (MAK) to automate the processes (http://perl.idmb.tamu.edu/mak.htm). In addition to its ability to routinely retrieve family member sequences and to report the positions of these elements relative to the closest neighboring genes, MAK is a powerful tool for revealing anchor elements that link MITE families to known transposable element families. Implementation of the MAK is described, as are genetic principles and algorithms used in its derivation. Test runs of the programs for several MITE families yielded anchor sequences that retain TIRs and coding regions reminiscent of transposases. These anchor sequences are consistent with previously reported putative autonomous elements for these MITE families. Furthermore, analysis of two MITE families with no known links to any transposon family revealed two novel transposon families, namely Math and Kid, belonging to the IS5/Harbinger/PIF superfamily.
Figures


References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions

