SVM-Prot: Web-based support vector machine software for functional classification of a protein from its primary sequence
- PMID: 12824396
- PMCID: PMC169006
- DOI: 10.1093/nar/gkg600
SVM-Prot: Web-based support vector machine software for functional classification of a protein from its primary sequence
Abstract
Prediction of protein function is of significance in studying biological processes. One approach for function prediction is to classify a protein into functional family. Support vector machine (SVM) is a useful method for such classification, which may involve proteins with diverse sequence distribution. We have developed a web-based software, SVMProt, for SVM classification of a protein into functional family from its primary sequence. SVMProt classification system is trained from representative proteins of a number of functional families and seed proteins of Pfam curated protein families. It currently covers 54 functional families and additional families will be added in the near future. The computed accuracy for protein family classification is found to be in the range of 69.1-99.6%. SVMProt shows a certain degree of capability for the classification of distantly related proteins and homologous proteins of different function and thus may be used as a protein function prediction tool that complements sequence alignment methods. SVMProt can be accessed at http://jing.cz3.nus.edu.sg/cgi-bin/svmprot.cgi.
Figures


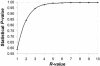
References
-
- Eisenberg D., Marcotte,C.A., Xenarios,I. and Yeates,T.O. (2000) Protein function in the post-genomic era. Nature, 405, 823–826. - PubMed
-
- Bork P., Dandekar,T., Diaz-Lazcoz,Y., Eisenhaber,F., Huynen,M. and Yuan,Y. (1998) Predicting function: from genomes and back. J. Mol. Biol., 283, 707–725. - PubMed
-
- Pellegrini M. (2001) Computational methods for protein function analysis. Curr. Opin. Chem. Biol., 5, 46–50. - PubMed
-
- Teichman S.A. and Mitchison,G. (2000) Computing protein function. Nat. Biotechnol., 18, 27. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

