PipeAlign: A new toolkit for protein family analysis
- PMID: 12824430
- PMCID: PMC168925
- DOI: 10.1093/nar/gkg518
PipeAlign: A new toolkit for protein family analysis
Abstract
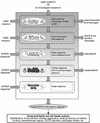
PipeAlign is a protein family analysis tool integrating a five step process ranging from the search for sequence homologues in protein and 3D structure databases to the definition of the hierarchical relationships within and between subfamilies. The complete, automatic pipeline takes a single sequence or a set of sequences as input and constructs a high-quality, validated MACS (multiple alignment of complete sequences) in which sequences are clustered into potential functional subgroups. For the more experienced user, the PipeAlign server also provides numerous options to run only a part of the analysis, with the possibility to modify the default parameters of each software module. For example, the user can choose to enter an existing multiple sequence alignment for refinement, validation and subsequent clustering of the sequences. The aim is to provide an interactive workbench for the validation, integration and presentation of a protein family, not only at the sequence level, but also at the structural and functional levels. PipeAlign is available at http://igbmc.u-strasbg.fr/PipeAlign/.
Figures
References
-
- Lecompte O., Thompson,J.D., Plewniak,F., Thierry,J. and Poch,O. (2001) Multiple alignment of complete sequences (MACS) in the post-genomic era. Gene, 270, 17–30. - PubMed
-
- Plewniak F., Thompson,J.D. and Poch,O. (2000) Ballast: blast post-processing based on locally conserved segments. Bioinformatics, 9, 750–759. - PubMed
-
- Thompson J.D., Plewniak,F., Thierry,J. and Poch,O. (2003) RASCAL: Rapid scanning and correction of multiple sequence alignment programs. Bioinformatics, in press. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous