G protein-coupled receptor drug discovery: implications from the crystal structure of rhodopsin
- PMID: 12825452
- PMCID: PMC1383658
G protein-coupled receptor drug discovery: implications from the crystal structure of rhodopsin
Abstract
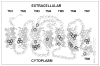
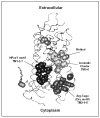
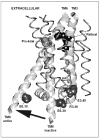
G protein-coupled receptors (GPCRs) are a functionally diverse group of membrane proteins that play a critical role in signal transduction. Because of the lack of a high-resolution structure, the heptahelical transmembrane bundle within the N-terminal extracellular and C-terminal intracellular region of these receptors has initially been modeled based on the high-resolution structure of bacterial retinal-binding protein, bacteriorhodopsin. However, the low-resolution structure of rhodopsin, a prototypical GPCR, revealed that there is a minor relationship between GPCRs and bacteriorhodopsins. The high-resolution crystal structure of the rhodopsin ground state and further refinements of the model provide the first structural information about the entire organization of the polypeptide chain and post-translational moieties. These studies provide a structural template for Family 1 GPCRs that has the potential to significantly improve structure-based approaches to GPCR drug discovery.
Figures




References
-
- Sautel M, Milligan G. Molecular manipulation of G-protein-coupled receptors: A new avenue into drug discovery. Curr Med Chem. 2000;7:889–896. - PubMed
-
- Muller G. Towards 3D structures of G protein-coupled receptors: A multidisciplinary approach. Curr Med Chem. 2000;7:861–888. ● This review summarizes the recent results on GPCR structural studies. - PubMed
-
- Bargmann CI. Neurobiology of the Caenorhabditis elegans genome. Science. 1998;282:2028–2033. - PubMed
-
- Larhammar D, Blomqvist AG, Wahlestedt C. The receptor revolution - multiplicity of G-protein-coupled receptors. Drug Des Discov. 1993;9:179–188. - PubMed
-
- Howard AD, McAllister G, Feighner SD, Liu Q, Nargund RP, Van der Ploeg LH, Patchett AA. Orphan G-protein-coupled receptors and natural ligand discovery. Trends Pharmacol Sci. 2001;22:132–140. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
